![]() Submit two
introductions - one of database and one of analysis tool.
Submit two
introductions - one of database and one of analysis tool.
http://www.ncbi.nlm.nih.gov/Genbank/index.html
GenBank is NCBI's primary sequence database. The data is shared nightly among
three collaborating databases, GenBank, DNA Database of Japan (DDBJ). Mishima,
Japan, European Molecular Biology Laboratory Database (EMBL) at EBI. Hinxton,
UK. It has bulk sequence divisions as follow:PAT Patent, EST Expressed Sequence
Tags (111 files), STS Sequence Tagged Site, GSS Genome Survey Sequence (37
files), HTG High Throughput Genome (25 files), HTC High Throughput cDNA, CON
Contig.
http://www.ncbi.nlm.nih.gov/BLAST/
BLAST is Basic Local Alignment Search Tool. It is a widely used similarity
search tool for homologs and a heuristic approach based on Smith Waterman
algorithm. It finds best local alignments and provides statistical significance.
It has all combinations (DNA/Protein) query and database: DNA vs DNA, DNA
translation vs Protein, Protein vs Protein, Protein vs DNA translation, DNA
translation vs DNA translation.
For further information, you can go to its website http://www.ncbi.nlm.nih.gov/
to look up.
![]() Make your own home page! Put your assignment 1 here too.
This will be the place for you to submit your future assignments.
Make your own home page! Put your assignment 1 here too.
This will be the place for you to submit your future assignments.
http://www.life.nthu.edu.tw/~b881605
![]() Search
genes associated with colon cancer in human genome.MLH1 was identified as a
locus frequently mutated in hereditary nonpolyposis colon cancer (HNPCC). When
cloned, it was discovered to be a human homolog of the E. coli mismatch repair
gene mutS.
Search
genes associated with colon cancer in human genome.MLH1 was identified as a
locus frequently mutated in hereditary nonpolyposis colon cancer (HNPCC). When
cloned, it was discovered to be a human homolog of the E. coli mismatch repair
gene mutS.
1. How many hits will you get if you search genes associated with colon cancer
in human genome?
2. How many loci will you find if you search locus link for human in Genebank?
3. Give the locus ID and position of MLH1.
Locus ID 4292 and position 3p21.3.
4. Find the %ID of nucleotide sequence for its possible orthologs in mouse.
| Human | Hs.57301 BC005866 LocusLink 4292 MLH1 |
90.5 | Mm.4438 AK010617 LocusLink 15361 Hmga1 |
Mouse |
5. Find the total number of mutations of MLH1 reported in human gene mutation
database.
6. Give the DNA sequence of MLH1.
7. Give the DNA sequence of E. coli mismatch repair gene mutS.
Click here to see.
![]() Compare
human colon cancer gene MLH1 with other genes.
Compare
human colon cancer gene MLH1 with other genes.
- To use ORF finder to translate DNA sequence to protein sequence in all reading
frames. -
- To use blastn, blastp, CD search and blast 2 sequence programs for searching
and comparison. -
1. Compare MLH1 (answer of assignment 3.6) and mutS (answer of 3.7) sequence.
¡@
Use "Blast 2 Sequences."
The result is "No significant similarity was found."
2. Translate the above two gene sequences to protein sequences.
¡@
If using "ORF finder" to translate, we would get results as below : MLH1 protein sequence. and mutS protein sequence. If using Genbank to find the product protein directly, we would get results as below : MLH1 protein sequence. and mutS protein sequence. The results are the same.
3.Perform protein sequence homology searching for MLH1 in GenBank. Give the 10
highest hits.
¡@
Use "blastP" in ORF finder.
4. Compare human MLH1 protein with MLH1 in M. musculus, R. norvegicus and D.
melanogaster. Give the pairwise alignment and % of sequence similarity.
Use the result for last question.
Mus musculus : Identities = 409/484 (84%), Positives = 439/484 (90%), Gaps = 4/484 (0%), pairwise alignment
Rattus norvegicus : Identities = 659/758 (86%), Positives = 707/758 (92%), Gaps = 3/758 (0%), pairwise alignment
Drosophila melanogaster : Identities = 345/751 (45%), Positives = 472/751 (61%), Gaps = 94/751 (12%), pairwise alignment
5. Search the conserve domain (CD) for MLH1. Give the position of the CD, name
of CD and Pfam ID number.
6. Show multiple alignment of MLH1 conserve domain with 5 sequences from the top
of the CD alignment
![]() Sequence
analysis of MLH1 protein.
Sequence
analysis of MLH1 protein.
- To search protein file in SWISS-PROT Database
- To use analysis tools in ExPASy
1. Search human MutL protein homolog 1, Mlh1, in SWISS-PROT database. Give its
(a) accession number, (b) entry name and (c) release date of last modification.
¡@
SWISS-PROT >> SRS >> enter"Mlh1" >> answer
(a) accession number : P40692
(b) entry name : MLH1_HUMAN
(c) release date of last modification : Release 31, February 1995
2. Give its number of amino acids, molecular weight and theoretical pI.
¡@
Number of amino acids : 756aa
Use "Compute pI/Mw" of "Tools" to get the answer.
Molecular weight : 84600.98
Theoretical pI : 5.51.
3. Calculate the total number of negatively charged residues and positively
charged residues.
¡@
Use "ProtParam" of "Tools" to get the answer.
Total number of negatively charged residues (Asp + Glu): 104
Total number of positively charged residues (Arg + Lys): 83
4. Calculate its hydrophobicity (Kyte & Doolittle scale, window size 11,
Relative weight 60%).
¡@
Use "ProtScale" of "Tools" to get the answer.
5. Performe the trypsin (higher specificity) cleavage of the protein. (a) How
many peptides will you get after cleavage? (b) Give the list of peptides with a
mass bigger than 1000 dalton.
¡@
Use "PeptideMass" of "Tools" to get the answer.
¡@
![]() Multiple
Sequence Alignment of MLH1 protein.
Multiple
Sequence Alignment of MLH1 protein.
- To do Multiple Sequence Alignment in Biology Workbench
- To draw phylogenetic tree from alignment
1. Add MLH1_Human protein to the Biology Workbench. Predict its secondary
structure by GOR4.
Biology
workbench >> Protein tools >> use
"Ndjinn (Multiple Database Search)" >> enter"MLH1"
>> use GENPEPT (GenBank Gene Products Last Full Release)
>> select "GENPEPT: 463989Human DNA mismatch
repair protein homolog (hMLH1) mRNA, complete" >> import sequence
>> use "GOR4 (Predict Secondary Structure of PS)" in protein
tools >> answer.
2. Do a homology searching of MLH1_Human in Genpept Full Release Database.
Import MLH1-like protein of C. elegans, S. cerevisiae, D. melanogaster, R.
norvegicus and M. musculus to your workbench. Run CLUSTALW to get multiple
sequence alignment for these six proteins.
use "FASTA-Heuristic Sequence Similarity Search (PS Or DB)" in protein tools >>
3. Perform BOXSHADE program to get a color-coded plot for the results of
question 2.
4. Draw rooted phylogenetic tree for these proteins.
answer
¡@
![]() Protein
Structure Database -PDB-.
Protein
Structure Database -PDB-.
- To do Protein Structure data search
- To understand protein structure data file
¡@
1. Search human Mlh1 or its homologous proteins in Protein Data Bank (PDB) database. You just need to search for protein structure, do not find complex structure. Give its (a) ID number, (b) Name of the molecule, (c) experimental method for structure determination and (d) Resolution of the structure.
PDB >> enter "mutL" (a homolog of Mlh1) >> select one that is not complex structure >> 1BKN
(a) Mol_Id: 1; Molecule: Mutl;
(b) N-Terminal 40Kd Fragment
(c) Exp. Method:X-ray Diffraction
(d) Resolution [Å]:2.90
2. Show its structure in ribbons form (400x400).
select "View Structure" in the left column >> Custom Size Images size : (400) >> answer
3. Give the structural informations of this PDB file (a) number of protein
chains, (b) number of helix in one protein chain, (c) the first and last
residues in the helix 6 .
select "Download/Display File" in the left column >> answer
(a) number of protein chains: 2
(b) number of helix in one protein chain: 11 helix in A chain and 10 helix in B chain
(c) the first and last residues in the helix 6: THR A 166 ALA A 181
4. Give the Class, Fold, Superfamily and Family of this protein in Structural
Classification of Proteins (SCOP).
select "Other Sources" in the left column >> SCOP (Structural Classification) >> answer
¡@
Protein: DNA mismatch repair protein MutL from Escherichia coli [d.122.1.2]
Protein: DNA mismatch repair protein MutL from Escherichia coli [d.14.1.3]
5. (a) Show Ramachandran plot for this PDB file. (b) How many residues are in
the most favoured regions of Ramachandran plot?
(a) select "Other Sources" in the left column >> Procheck (Strutcure Summary) >> Ramachandran plot >> answer
(b) 379 residues.
¡@
![]() Using
Molecular Viewer to see the features of Protein Structure
Using
Molecular Viewer to see the features of Protein Structure
- To familiar with RasMol
- To understand protein structure features by RasMol commands
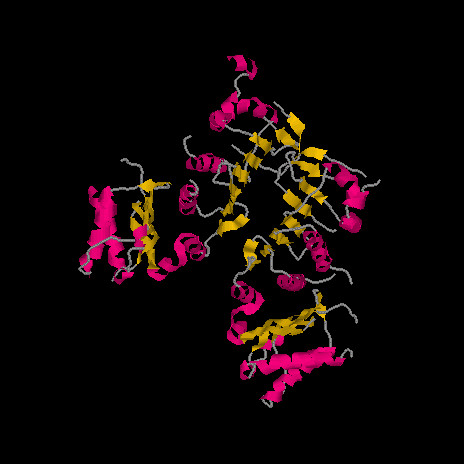
1. Make a GIF picture of the PDB file that you find in assignment 7 by RasMol.
Display it in cartoon form and color it by structure. Please make the
orientation of the molecule just like the molecule in the picture of assignment
7 - 2.
¡@
¡@
PDB >> enter "1BKN" >> 1BKN >> view structure >> Rasmol >> get 1BKN Rasmol file
¡@
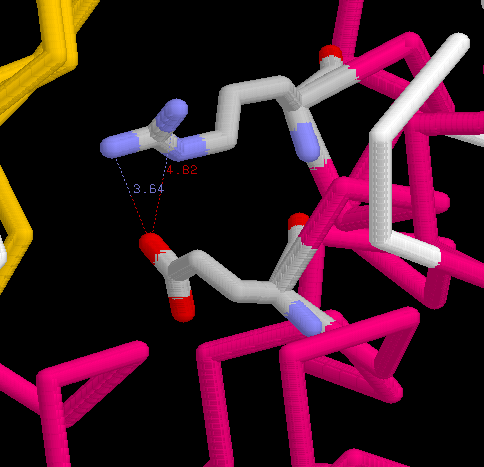
2. Residues Glu173 and Arg177 are possible to form an ion pair (salt-bridge).
(a) Draw a zoom-in picture of this pair. Show these two residues in sticks and
color with cpk. The rest of protein in backbone form. (b) Measure the distance
between Glu173A.OE and ARG177A.NH.
¡@
¡@
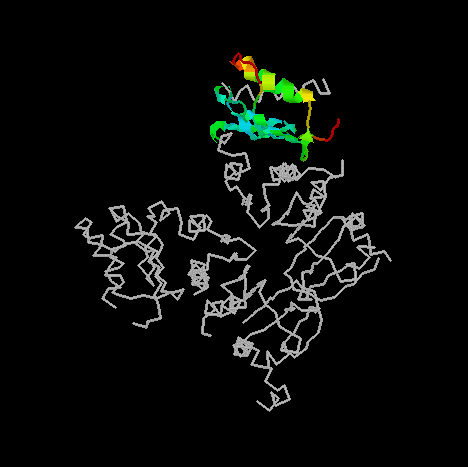
3. Show a protein fragment with residues 622 to 699 only. Show them in cartoon form and color with temperature.
¡@
4. Make a stereo picture for only the atoms within 6.0 Angstrom of Phe721. Show
this residue in green color, others in cpk and all the residues in wireframe 0.1
format.
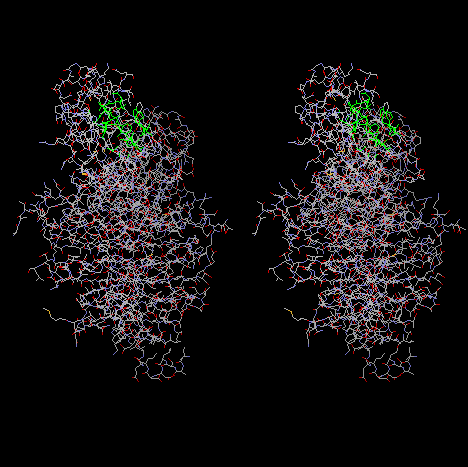
¡@
![]() Comparative
Protein Modelling by SWISS-MODEL
Comparative
Protein Modelling by SWISS-MODEL
- To make a protein model by homology modelling
1. Make a protein structure model of MLH1 based on the protein you find in
assignment 7. Show their pairwise alignment.
¡@
2. How large is the fragment of MLH1 possible to be modeled?
¡@
336 residues (residues 3 - 339 of submitted sequence).
3. Show three rasmol pictures of your model of MLH1?
¡@