
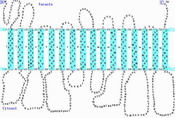
Vacuoles occupy 90% volume of plant cells. Plant vacuoles play an important role in the maintenance and turgor pressure and storage of ions and metabolites. Vacuolar membrane of plant cell contains two types of H+-pumps, a vacuolar H+-ATPase (EC 3.6.1.3) and a vacuolar H+-pyrophosphatase (PPase; EC 3.6.1.1), using ATP and pyrophosphate as energy sources, respectively. Both proton pumping system catalyze electrogenic H+ translocation from cytosol to the vacuole lumen to generate an inside-acidic and inside-positive membrane potential for the secondary transport of various ions and metabolites. Vacuolar H+-ATPase is ubiquitous proton pump in all eukaryotes, while vacuolar H+-PPase is found primarily in higher plants and a few species of phototropic bacteria, notably purple nonsulfur bacterium, and hyperthermophilic archaeon. Furthermore, many organelles contain different types of proton pumping ATPases; albeit, proton translocating PPase is generally observed on vacuole membrane.
Both proton pumping enzymes, residing on the same membrane of plant vacuoles, are functionally similar, but structurally distinct. Accordingly, we are interested in several issues: (1) Is possible that H+-PPase exist on other organelles? (2) What is the relationship between their structure and function? (3) How are both proton pumps regulated under physiological and stress conditions?
Exploration on the plant DNA end-binding proteins
Many environmental stress could cause the breakage, deletion, translocation, and deformation of chromosomes, threatening survival of organisms and possibly also to their offspring. To repair such damages, eukaryotes employ two following mechanisms: (1). homologous recombination pathway (HR), and (2). Non-homologous end joining pathway (NHEJ). A DNA-dependent protein kinase is found to be involved in the non-homologous end-joining pathway. DNA-dependent protein kinase is a sole protein kinase absolutely depending on the presence of DNA and contains a catalytic subunit, named DNA-dependent protein kinase catalytic subunit (DNA-PKcs) and a heterodimeric protein complex, named Ku-protein. Ku-protein is composed of two subunits with molecular masses of 70 (Ku-70) and 80 (Ku-80) kDa, respectively. DNA-PKcs recruits Ku-protein, and as Ku-protein binds DNA, DNA-PKcs phosphorylates other proteins associated on DNA for the repair of DNA breaks or for other possible physiological functions.
Besides, Ku-protein is involved in V(D)J recombination in immune cells. Recently, Ku-protein was found to play a role in maintaining the structure of telomere. Ku-deficiency results in cell arrest in G2 phase and prolong in cell cycle, suggesting the involvement of Ku-protein in the regulating cell growth and cell cycle. Several lines of evidence also indicate its role in regulation of replication, transcription, and cell signaling. Taken together, it is fascinatingly implicated Ku-protein belongs to a protein family of multifunction.

NanoBiotechnology
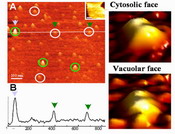
To understand how biomolecules are assembled, function, and work in situ, in vitro, and in vivo is of fundamental importance to investigate and manipulate them in their scale under native environments. Bionanotechnology is an emerging inter-discipline area of science and technology. Bionanotechnology, in one way, applies the tools and processes of nano/microfabrication to develop devices for studying biosystems in nanoscale size. On the other hand, workers also learn from biosystems how to prepare better nanoscale devices. This innovative integrated proposal is based on the most recent leading advances in the development of several new techniques and the achievements by this research team. This project then aims at employing a novel bionanotechnique combining the power of 3-D (3-dimensional) nanofabrication, nanofluid-assisted atomic force microscopy (AFM), BioMEMS (Bio-microelectromechanical systems), molecular bio-techniques, modern genetic and biochemical technologies altogether to investigate and manipulate single membrane protein H+-PPase, for understanding its basic structure, physiological functions, reaction mechanism, proton pumping efficiency, the role of intermolecular forces relating to structure variations under in situ, in vitro or in vivo conditions, and the interaction between H+-PPase and its surrounding membrane. In this proposal, genetically modified H+-PPase, in purified form or on cell membrane, will be brought into a nanohole region by electrowetting technique and self-assembled with artificial bilayer lipid membranes (BLMs) across the nano/microholes for registration. Nanofluid-assisted AFM cantilever tip will be modified in situ with designed biomolecules by the electrowetting-operated nanodroplet dispensing system. The investigation on dynamics of protein structure will be performed by nanofluid-assisted AFM cantilever together with nanoelectrode measurement of membrane potential on different sides in nanoscale for understanding real time proton-pumping efficiency of H+-PPase and its structure-function relationship especially in a dynamic state. Accordingly, the function and structure of H+-PPase can be monitored lively in molecular resolution by image, intermolecular force measurement as well as manipulation, and electrical detection in situ.
Bioinformatics
Elucidating
transcriptional regulation in plant genes is one of the most important
and urgent areas of research for plant scientists, following the
mapping of various plant genomes, such as A. thaliana, O. sativa and Z. mays.
A variety of bioinformatic servers or databases of plant promoters have
been established, although most of them have been aimed only at
annotating transcription factor binding sites in a single gene and have
neglected some important regulatory elements (tandem repeats and
CpG/CpNpG islands) on promoter regions. Additionally, the combinatorial
interaction of transcription factors (TFs) is important for regulating
the gene group that is associated with same expression pattern.
Therefore, a tool for detecting co-regulation of transcription factors
in a group of gene promoters is required.
This study develops a database-assisted system, PlantPAN (Plant Promoter Analysis Navigator), for recognizing combinatorial cis-regulatory
elements with distance constraint in plant co-expressed genes. The
system collects the plant transcription factor binding profiles from
PLACE, TRANSFAC public release 7.0, AGRIS, and JASPER databases and
allows users to input a group of gene IDs or promoter sequences,
enabling the co-occurrence of combinatorial transcription factor
binding sites (TFBSs) within a defined distance (20 bp to 200 bp) to be
identified. Additionally, the new resource enables the annotation of
other regulatory features in a plant promoter, such as CpG/CpNpG
islands and tandem repeats. Moreover, the regulatory elements in the
conserved regions of the promoters across homologous genes are detected
and displayed.
The PlantPAN not only provides a user-friendly input/output interface, but also has numerous advantages in analyzing a plant promoter. Several case studies demonstrate the effectiveness of PlantPAN. This novel analytical resource is now freely available at http://PlantPAN.mbc.nctu.edu.tw.
