GFP assay (yeast two hybrid )
Introduction
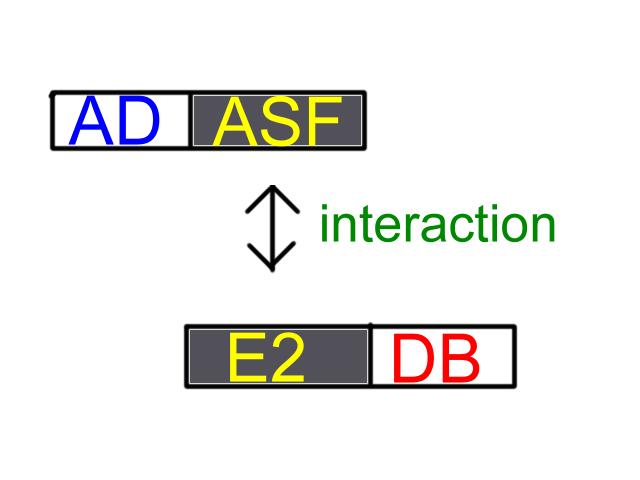
The two-hybrid system exploit thi ability of a pair of interacting
proteins to bring a transcription activation domain into close proximity
with DNA-binding site that regulates the expression of an adjacent
reporter gene. Generally the assay is performed in yeast. It requires the
construction of hybrid genes to encode:
(1) a DNA-binging domain(DB) fused to a protein A (in our case:E2)
(2) an activation domain(AD) fused to a protei B(in our case:ASF)
The domains most commonly used are the DNA-binding domains of
GAL4 and LexA, and the activation domain of Gal4 and Herpes virus VP16.
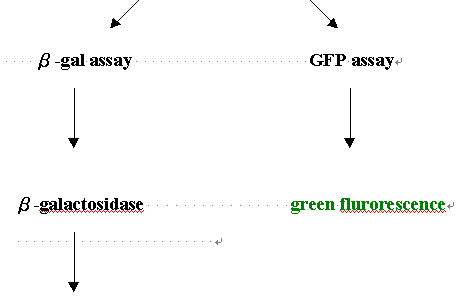
One reporter gene often used is E. coli lacZ, which produces blue colonies
on plates or filters containing X-gal. In addition, use of a yeast gene involved
in amino acid biosynthesis ,such as HIS3 or LEU2, allows selsction for cells
that grow on media lacking the revelant amino acid, and is particularly helpful.
We used the GFP repoter gene instead of lacz to see if the colonoes will
produce fluroscence proteins and can be visualize under UV light.
Method
It is known that the hinge of E2 protein interacts with the alternative
splicing factor, so we set 6 reactions:
|
DB
|
AD
|
LacZ/GFP
|
|
empty
|
ASF
|
|
E2(full-length)
|
ASF
|
|
E2กต2(truncated):hinge(RS
domain) is deleted
|
ASF
|
ASF(Alternative splicing factor)
 Result
The E2-ASF set is not very distinguishable from the empty-ASF
set under UV light, probably due to:
(1) too late to examine the colonies under UV light;
(2) the basal level transcription in empty-ASF colony
Reference:
The two-hybrid system: an assay for protein-protein interactions
STALEY FIELD AND ROLF STERNGLANZ
TIG AUGUST 1994 Vol.10 No8
Result
The E2-ASF set is not very distinguishable from the empty-ASF
set under UV light, probably due to:
(1) too late to examine the colonies under UV light;
(2) the basal level transcription in empty-ASF colony
Reference:
The two-hybrid system: an assay for protein-protein interactions
STALEY FIELD AND ROLF STERNGLANZ
TIG AUGUST 1994 Vol.10 No8
 to abstract
to abstract

 Result
The E2-ASF set is not very distinguishable from the empty-ASF
set under UV light, probably due to:
(1) too late to examine the colonies under UV light;
(2) the basal level transcription in empty-ASF colony
Reference:
The two-hybrid system: an assay for protein-protein interactions
STALEY FIELD AND ROLF STERNGLANZ
TIG AUGUST 1994 Vol.10 No8
Result
The E2-ASF set is not very distinguishable from the empty-ASF
set under UV light, probably due to:
(1) too late to examine the colonies under UV light;
(2) the basal level transcription in empty-ASF colony
Reference:
The two-hybrid system: an assay for protein-protein interactions
STALEY FIELD AND ROLF STERNGLANZ
TIG AUGUST 1994 Vol.10 No8
 to abstract
to abstract