Advance Molecular Biology
(LS6421, 2002) Part
IV-3 T.
Y. Lin
Regulation of transcription
1.
The
level of control in the expression of genes
(1). Activation
of gene structure
(2). Initiation
of transcription
(3). Processing
of the transcript
(4). Transport
to cytoplasm
(5). Translation
of mRNA
2.
Response elements identify genes under common regulation.
(1). Response
elements may be located in promoters (such as an HSE) or enhancers (such as a
GRE).
(2). Heat shock
transcription factors (HSTFs) and HSEs
(3). The regulatory region of a human
metallothionein (MT) gene contains constitutive elements in its promoter and
enhancer.
(4). The response to steroid hormone is
regulated by a GRE (at ~ -250 bp).
3.
Types of DNA-binding domains
(1). Proteins
regulate transcription by using particular motifs to bind DNA.
DNA-binding
Domains: protein motifs that are involved with binding to DNA
structures found principally in the major groove.
a.
The zinc finger motif (TFIIIA,
steroid receptors)
c.
The helix-turn-helix motif (phage repressors, homeodomain, mammalian transcription
factors).
d. The amphipathic
helix-loop-helix motif (developmental regulators) enables proteins to
dimerize, and a basic region near these motif contacts DNA.
e.
The leucine zippers form a dimmer. A stretch of positive charge residues is
involved in binding to DNA.
(2). Regulation of the activity of an inducible transcription factor.
a.
Synthesis of
protein (tissue-specific, homeodomain proteins)
b.
Covalent modification
of protein (HSTF is converted to active form by phosphorylation; AP1 (Jun/Fos heterodimer) by phosphorylating Jun)
c.
Ligand binding
(steroid receptors): activated or inactivated.
d.
Cleavage to release active factor (absence of sterol response)
e.
NF-kB is
sequestered in the cytoplasm by inhibitory protein I-kB.
In B-lymphocytes, NF-kB is released from
I-kB and moves to nucleus.
f.
Change of partner (HLH, MyoD/ID)
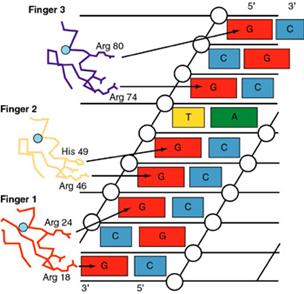
4. The zinc-finger
(Cys2/His2) motif
The zinc finger domain is formed by the interaction of
one or in some cases two Zn atoms with regions of the protein. The "finger" points into the
major groove.

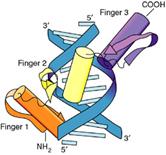
(1). A common motif in DNA-binding proteins such as SP1.
a.
The
consensus sequence of a single finger (classic) is Cys-X2-4-Cys-X3-Phe-X5-Leu-X2-His-X3-His
b.
The fingers usually are organized as a single series of
tandem repeats.
c.
The
loops of amino acids protrude from the zinc-binding site.
(2). Steroids
receptors (glucocorticoid and estrogen receptors)
a.
The Cys2/Cys2 finger consensus is Cys-X2-Cys-X13-Cys-X2-Cys
b.
Proteins
with Cys2/Cys2 fingers
often have nonrepeatitive fingers.
c.
Binding
sites in DNA are short and palindromic.
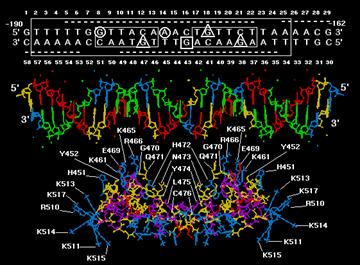
d.
One
side of the N-terminal helix makes contacts in
the major groove of DNA. Two glucocorticoid
receptors dimerize upon binding to DNA.
f.
Glucocorticoids
regulate gene transcription by causing their receptor to bind to an enhancer (glucocorticoid response element).
5.
Steroid receptors
have several independent domains.
(1). The regions include an individual N-terminal region (least conserved), conserved DNA-binding region, and a C-terminal
hormone-binding region.
(2). The C-terminal
hormone-binding region regulates the activity of the receptor in a way
that varies for the individual receptor.
a.
The glucocorticoid receptor:
If the C-terminal is deleted, the remaining N-terminal
protein is constitutively active. Þ In the absence of steroid, the
steroid-binding domain functions as an internal
negative regulator.
b.
The estrogen receptor:
If the hormone-binding
domain is deleted, the protein is unable to activate transcription, although it
continuously binds to the ERE.
(3). The receptor binds as a multimer.
(4). The response elements may be palindromes or
direct repeats.
(5). The recognition of response elements by a variety of receptors
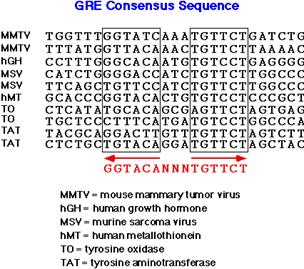
a. Glucocorticoid (GR),
mineralocorticoid (MR), androgen (AR) and progesterone (PR) receptors form homodimers with consensus sequence of half sites (RE: TGTTCT,
ER: TGACCT) that are arranged as palindromes; spacing between the sites
determines type of element.
b. Tyroid (T3R), vitamin D (VDR),
retinoic acid (RAR) and 9-cis-retinoic acid (RXR)
receptors form heterodimers, which recognize
half element TGACCT arranged as direct
repeats and recognition influenced by separation: 1 bp – RXR; 3 bp – VDR; 4 bp
- T3R; 5 bp – RAR
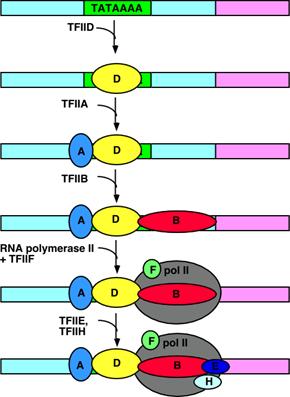
Review the transcription of a series of
yeast galactose (GAL) genes
Yeast expressed genes necessary to utilize galactose
when the sugar is present. The
transcription of these genes is induced by galactose.
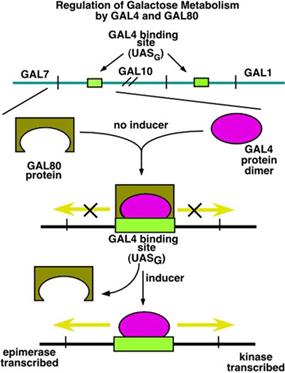
The GAL genes
are controlled from an upstream activating sequence (UAS). In the case of the one shown between GAL7
(galactose epimerase) and GAL10 (galactose kinase), transcription is regulated
in two directions. In the absence of galactose (the inducer) two
proteins bind to the UAS: the GAL4 protein (as
a dimer) and GAL80. GAL80
acts to block the transcription activation of GAL4.
When galactose
is present, GAL80 is removed. In some sense this is like the removal of
lac repressor from the operator. However,
the removal of GAL80 by itself does not activate the
transcription. It is the
presence of the GAL4 protein that causes transcription to increase.
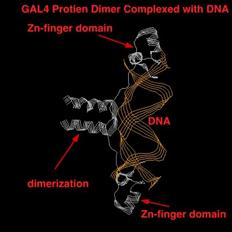
The GAL4 Zn-finger
domain activator works to increase transcription by a process called
recruitment. Recruitment means that the activator, once bound to
the binding site, causes the polymerase complex (pol II and the TFII's) to bind
more efficiently or more often to the promoter, thus increasing the rate of
transcription.
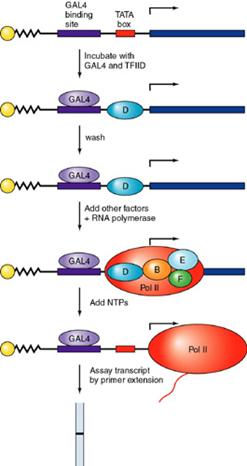
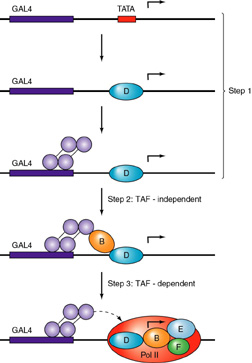
It can be
seen that GAL4 recruits the polymerase complex by interaction with TFIIB
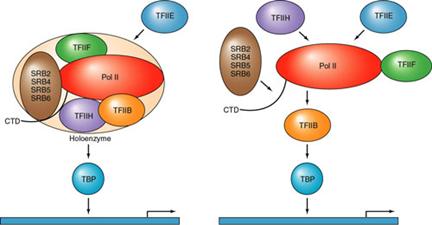
Two models are presented: on the
left, the recruitment of a holoenzyme complex
and on the right the recruitment, in sequence, of the
individual TFII's and the polymerase. (SRB 2, 4, 5, and 6 are protein
co-factors identified in a preparation of the enzyme that is called the
holoenzyme)
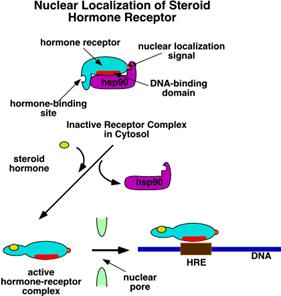
The Steroid Hormone Receptor
The general pattern of action, typified by glucocorticoids,
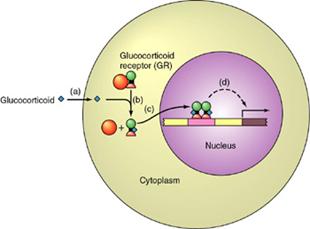
The hormone is hydrophobic and so
must arrive at the target cell via a carrier molecule. At the cell surface it is able to diffuse
across the lipid bilayer. In the
cytoplasm it must be taken up by a receptor.
The receptor
protein is found in an inactive form in the cytoplasm, complexed with hsp90 (orange
ball). When the hormone
binds, the receptor is activated and immediately moves to the nucleus. There, the Zn-finger domains bind to
sites on the DNA called hormone receptor elements
(HRE's) and gene expression is activated.
The
general structure of the hormone receptors is shown here:
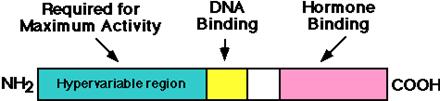
The addition of hormone causes the receptor to become a
transcriptional activator
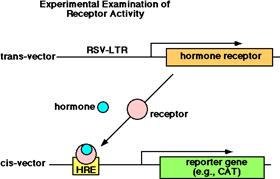
The trans-vector has the gene for the hormone receptor located downstream from a
very strong promoter (RSV-LTR). The
cis-vector has the hormone
response element upstream from a reporter gene. Place both vectors in the cell and then
add the hormone.
To make hybrid proteins using
this system: make a trans-vector with the DNA binding
domain of a glucocorticoid receptor and the hormone
binding domain of an estrogen receptor. This allows you to study the
interaction of hormone, receptor and HRE.
6.
Homeodomains bind related targets in DNA.
Homeodomain regions are found in proteins such as those
responsible for regulated development in Drosophila.
These are essentially
helix-turn-helix motifs. 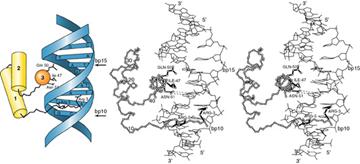
(1). The homeodomain (60 residues) may be the sole DNA-binding motif in a transcriptional regulator or may
be combined with other motifs (pou region).
(2). The homeodomain starts with the N-terminal arm, and the three
helical regions occupy residues 10-22, 28-38 and 42-58.
(3). Helix
3 of the homeodomain binds in the major groove of DNA and contacts both the
phosphate backbone and specific bases. Helices 1 and 2 lay outside the double
helix. N-terminal
arm lies in the minor groove.
7.
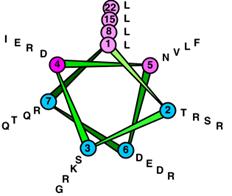
Helix-loop-helix proteins (40-50 aa) interact by combinatorial
association.
(1). All HLH proteins have
two amphipathic helices (with the ability to form dimer) separated
by a loop (10-24 aa).
(2). Basic HLH has a
region with positive charges adjacent to helix
1.
a.
Class A consists
of proteins that are ubiquitously expressed
(mammalian E12/E47; Drosophilae
da).
b.
Class B consists
of proteins that are expressed in a tissue-specific
manner (mammalian MyoD, myogenin, Myf-5; Drosophilae
Ac-S).
(3). Dimers formed from bHLH proteins
differ in their abilities to bind to DNA.
a.
E47 homodimers, E12-E47 heterodimers and MyoD-E47 heterodimers all
form efficiently and bind strongly to DNA.
b.
E12 homodimerizes well but binds DNA poorly, while MyoD
homodimerizes only poorly.
c.
E12 possesses an inhibitory region just by the basic region, which
prevents DNA binding by homodimers.
(4). The
distinction between the nonbasic HLH and bHLH proteins
a.
bHLH proteins (such as AC-S and da) dimerize and bind DNA
b.
HLH
proteins that lack the basic region (emc and Id) prevent DNA-binding.
c.
The trigger for muscle differentiation may be a heterodimer
consisting of MyoD-E12 or MyoD-E47.
Before myogenesis, Id may bind to MyoD, E12 or E47 to form heterodimers
that cannot bind to DNA.
The helix-loop-helix is a
dual function domain. The
helix-loop-helix region is the dimerization domain and is a set of two
amphipathic helices. The basic
region is the DNA binding domain.
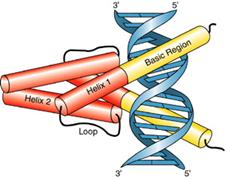
8.
Leucine zippers are involved in dimmer formation
(1). The basic regions of
the bZIP motif are held together by the dimerization at the adjacent
zipper region when the hydrophobic faces
of two leucine zippers interact in parallel orientation.
(2). The basic regions
bifurcate symmetrically to form arms that bind to DNA.
(3). Zippers may be used to sponsor formation of
homo- or heterodimers.
(4). Leucine occupies every seventh residue in the
potential zipper (4 repeats in C/EBP and 5 repeats in
Jun and Fos (AP1)).
The leucine zipper
is an amphipathic helix.
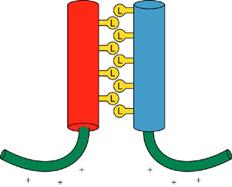
The protein functions only
when the dimer is formed. Dimer
formation is driven by the hydrophobic effect, as water increases in entropy
when the leucine faces are together.
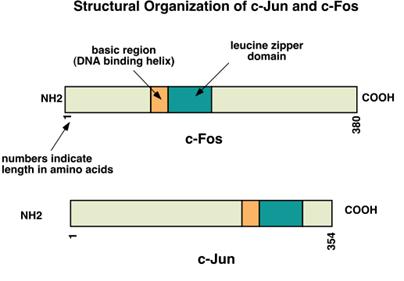
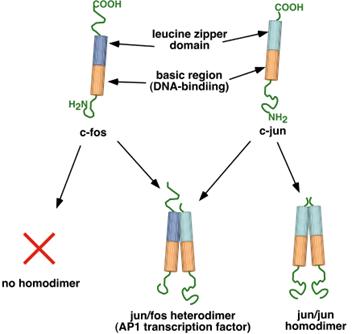
Jun and fos proteins are both of the basic helix/leucine zipper
type (bZIP). Fos is a protein found
in the Finkel-Biskis-Jenkins murine osteosarcoma virus. Jun is a protein first identified by
Japanese workers as a gene in the avian sarcoma virus 17.
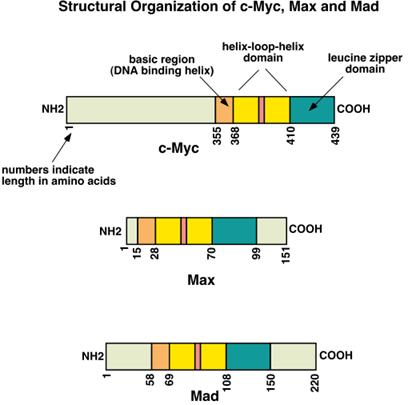
myc- Family
The myc family contains the bHLH (helix-loop-helix) motif. Myc itself was discovered as a protein of
the avian myelocytoma virus. Another member of the family is Max, which stands
for "myc activation substance X." A third member of this family is called
Mad, standing for "max dimerizer".
These proteins contain both the HLH and the
leucine zipper domains, along with the basic region that binds to DNA.
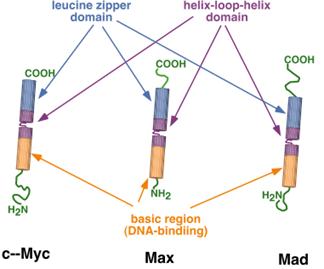
This family is a very powerful set of
transcriptional activators and repressors, active all throughout early
mammalian development. They
interact with a very specific DNA sequence: CACGTG. All sorts of homo- and heterodimers are
possible with this group.
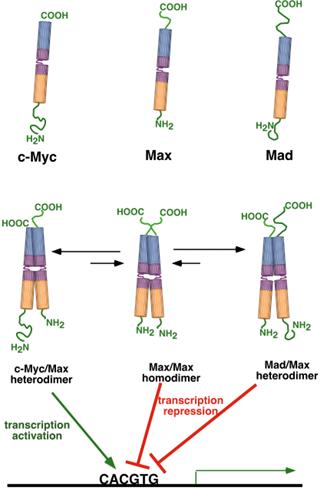
The combinations Max/Max and Mad/Max
function as repressors of transcription, while the Myc/Max is a strong
activator. When Myc is present, the
equilibrium is shifted to Myc/Max dimers, but when Mad is present, the
equilibrium is shifter to Mad/Max dimers. In this way, the formation of the
heterodimers controls the relative rate of transcription from genes controlled
by this family.
9.
Chromatin
remodeling is an active process
(1). The pre-emptive model
a.
If
nucleosomes form at a promoter, transcription factors cannot bind.
b.
Competition exists
between histones and transcription factors.
c.
TFIIIA can form
the necessary complex with free DNA.
d.
If
histones are added before TFIID, transcription cannot be initiated. TFIID recognizes free DNA, but can’t recognize or function
on nucleosomal DNA.
(2). The dynamic model
a.
Transcription
factors can use energy provided by hydrolysis of ATP.
b.
GAGA transcription factor disrupts nucleosomes at its binding site even when added after
assembly of nucleosomes.
c.
The PHO system
(a). At
the PHO5 promoter, the bHLH regulator PHO4 responds to phosphate starvation by
inducing the disruption of four precisely positioned nucleosomes
(independent of transcription and replication).
(b). The
two binding sites for PHO4 at the PHO5 promoter,
one located between nucleosomes, which can be bound by the isolated DNA-binding domain of PHO4,
and the other within a nucleosome, which cannot
be recognized.
(c). Disruption of the nucleosome to allow DNA binding at
the second site is necessary for gene activation.
(d). Activator sequence of VP16
can substitute for that of PHO4 in nucleosome
disruption. Disruption occurs by
protein-protein interactions that involve the same region that makes
protein-protein contacts to activate transcription.
Transcription-activiating
Domains: three kinds of
protein domains that are involved in transcription activation.
1. acidic domains, where the
amino acid side chains are acidic in nature (glutamic acid, aspartic acid)
2. glutamine-rich domains,
with about 25% glutamine in the sequence
3. proline-rich domains
These domains interact with
components of the transcription complex.
(3). Interactions
between TFs and chromatin are required for activation.
a.
The mouse mammalian tumor virus (MMTV) promoter
(a). It
contains an array of 6 partly palindromic sites,
each bound by one dimer of hormone receptor (HR).
(b). It
has a single binding site for NF1 and two adjacent
sites for OTF.
(c). HR
is binding to DNA on the nucleosomal surface.
(c). After
hormone induction, the changes in nucleosomal structure thus allow NF1 to bind
and activate transcription.
(d). NF1
can be footprint on the nucleoside after hormone
induction.
b.
The SWI/SNF complex
(a). The SWI/SNF
complex comprises ~10 proteins with a MW of ~2x106.
(b). It has an ATPase activity (SWI2).
(c). The basic
role of the SWI/SNF complex is chromatin remodeling.
(d). The SWI/SNF complex stimulates binding of GAL4 to its target
site on nucleosomal DNA in vitro
(ATP-dependent).
10.
Histone acetylation and deacetylation control chromatin activity
(1). Histone
acetyltransferases (HATs) and histone deacetylases (HDACs)
(2). Group A of HAT is involved in transcription;
Group B is involved with nucleosome assembly.
(3). Trichostatin
and butyric acid inhibit histone deacetylases.
(4). The
catalytic subunit of a group A HAT was identified as a homologue of the yeast
regulator protein GCN5. GCN5 has
HAT activity on H3 and H4.
(5). p300 p300.doc/CBPCBP02.doc is a coactivator that links
an upstream TF to the basal apparatus.
a.
The
p300/CBP interacts with various TFs, including hormone receptors, AP-1 and
MyoD. The interaction is inhibited
by viral regulator proteins adenovirus E1A and SV40 T antigen.
b.
The p300/CBP
acetylates the N-terminal tails of H4 in nucleosomes.
c.
PCAF, another coactivator, preferentially acetylates H3 in
nucleosomes.
d.
The
presence of multiple HAT activities in a coactivating complex: each HAT has a
different specificity.
(6). Drosophila
TAFII250 (ubiquitin-activating/conjugating)
binds to the acetylated tails of core histones throught its bromodomains. This interaction may enable TAFII250
to ubiquitinate H1 thus altering the accessibility of chromatin to TFs.
(7). Deacetylation: a repressor complex contains three
components: a DNA binding subunit, a corepressor, and a histone deacetylase.
a.
Yeast
Rpd3 has histone deacetylase activity.
b.
SIN3 (corepressor) & Rpd3 form complex with
DNA-binding protein Ume6.
11. Polycomb and trithorax are antagonistic repressors and
activators.
(1). Chromatin can be specifically repressed.
a.
Heterochromatin.
b.
Polycomb group (Pc-G)
represses homeotic genes.
(a). Pc is a nuclear protein (~80 sites on polytene chromosomes).
(b). These gene products form a general repressive complex that is
modified by some of the others for specific loci.
(c). Pc-G proteins do not initiate
repression, but are responsible for maintaining.
(d). If
Pc-G proteins are absent, the gene becomes activated.
(2). The
Polycomb response element (PRE) is 10 kb.
a.
No individual member of the Pc-G proteins has yet
been shown to bind to specific sequences in PRE.
b.
When Pc-G proteins repress a locus, the proteins
appear to be present over a large length of DNA than the PRE itself.
(3). A connection between the Pc-G complex and more general
structural changes in chromatin.
a.
A homology (chromodomain)
between a 37 amino acid region near the N-terminus of Pc and a nonhistone
protein, HP1, that is associated with heterochromatin.
b.
HP1 is coded by a gene Su(var)205, a suppressor of position-effect
variegation. Chromodomain may be used
to interact with common components that are involved in inducing the formation
of heterochromatin or inactive structures.
(4). The trithorax group (trxG) of proteins
a.
They act to maintain genes in an active state.
b.
The sites where Pc-G binds to DNA coincide with the
sites where GAGA factor (trithorax-like) binds
12.
Long range regulation and insulation of domains
(1). The human b-globin gene
a.
The 5'
regulatory sites are the primary regulators, and the cluster of hypersensitive
sites is called the LCR (locus
control region).
b.
Transfecting
various constructs into mouse erythroleukemia cells shows that the removal of
the LCR reduces the overall level of expression.
c.
LCR
may be required to open up the whole domain for transcription.
(2). The insulators
a.
When
an insulator is placed between an enhancer and a promoter, it prevents the enhancer from activating the promoter.
b.
Specialized
chromatin structures (scs (350 bp)
and scs' (200 bp))
(a). A region highly resistant to degradation of Dane I flanked on either
side by hypersensitive sites spaced at about 100 bp.
(b). The scs units do not play positive or negative roles
in controlling gene expression, but just restrict
effects from passing from one region to the next.
(c). Insulators block expression of any enhances that it separated from
the promoter.
(d). Mutations in
Su (Hw) abolish insulation. The Su (Hw)
gene codes for a protein that recognizes the insulator and is necessary for its
action. The insulator contains 12 binding sites for Su (Hw).
(e). Binding of Su (Hw) to DNA, followed
by binding of mod (mdg4) to Su (Hw), creates a unidirectional block to
activation of a promoter.
(f). The mod (mdg4) locus imposes
directionality on the ability of Su (Hw) to insulate promoters from the
boundary.
(3). Elements with different cis-acting properties are combined to
generate regions with complex regulatory effects.
a.
The Fab-7
region is a boundary element that is necessary for the independence
of regulatory elements iab-6 and iab-7.
b.
The
regulatory elements iab-6 and iab-7 control expression of the
adjacent gene Abd-B in successive regions of the embryo (segments A6 and
A7).
c.
Fab-7 may provide a boundary that prevents iab-7
from acting when iab-6 is usually active.
d.
Two
kinds of elements in the Fab-7
region: a sequence (~3.3 kb) behaves as an insulator
and sequence (~0.8 kb) behaves as a repressor that
acts on iab-7.
(4). There may be some sort of competitive
effect, in which the strength of the element determines how far its effects can
stretch.
(5). A possible chromosomal
domain
13.
Gene expression is associated with demethylation.
(1). A
majority of sites are methylated in tissues in which the gene is not
expressed. Demethylation is
required for gene expression.
(2). Nucleosomes
at the CpG islands have a reduced content of H1 histone, the other histones are
extensively acetylated and there are hypersensitive sites.
(3). House
keeping genes and CpG islands
(4). Repression
is caused by binding of MeCP-1 (several methyl groups are required) or MeCP-2
(a single methyl group) to methylated CpG sequences.
(5). MeCP-2,
which directly represses transcription by interacting with complex at the
promoter, is bound also to the Sin3 repressor complex, which contains histone
deacetylase activities.
(6). Gene
expression in the Diptern insects
(7). Methylation is
responsible for imprinting
a.
A
difference in behavior between the alleles inherited from each parent.
b.
The IGF-II gene of oocytes is methylated (silent), but
the IGF-II gene of sperm is not methylated (expressed).
14.
Pituitary-specific
POU domain factor Pit-1 activates growth hormone gene
expression in somatotrope cells. It
is achieved by actively repressing its expression in lactotropes in a manner
dependent on the presence of a single conserved Pit-1 recognition site.
Lawrence
Annu. Rev. Biochem. 2000.
69:729–49 MEDIATOR OF TRANSCRIPTIONAL REGULATION
Mediator complex: a conserved interface
between gene-specific regulatory proteins and the general transcription
apparatus of eukaryotes. Mediator
evidently integrates and transduces positive and negative regulatory
information from enhancers and operators to promoters.
DISCOVERY OF MEDIATOR
The products of four dominant
suppressors, termed Srb2, Srb4, Srb5, and Srb6, were shown to interact in a
large complex and bind to the polymerase CTD. The isolation of Mediator depended on
the complete resolution of all the general transcription factors. TFIIH fractions were contaminated with
Mediator. Mutational analysis of MED2
and MED6 has established their involvement in transcriptional regulation
in vivo. Early work showed that SRB2
functions through an upstream activating sequence (UAS) at the INO1 promoter,
and more recent studies have demonstrated MED2 function through a GAL
UAS. Deletion of the
nonessential MED2 gene caused a similar impairment of transcriptional
activation in vitro and in vivo. In
addition to enabling activated transcription, Mediator stimulates basal
transcription about 10-fold and stimulates phosphorylation of the polymerase
CTD by TFIIH kinase 30- to 50-fold.
The effect on basal transcription may relate to an apparent requirement
of Srb2 and Srb5 for transcription in a nuclear extract.