¡@
¡@
¡@
The enzyme N-carbamoylsarcosine amidohydrolase (CSHase; EC 3.5.1.59) hydrolyzes N-carbamoylsarcosine to sarcosine, carbon dioxide and ammonia. CSHase is involved in one of the two alternative pathways for creatinine degradation in microorganisms. The enzymes of the " creatine pathway" catalyze the degradation of creatinine to glycine via creatine and sarcosine , whereas in the CSHase-containing " N-methylhydantoin route" , glycine is produced by the degradation of creatinine with N-methylhydantoin, N-carbamoylsarcosine and sarcosine as intermediates. The enzymes of the N-methylhydantoin route (including CSHase) are the basis for the enzymatic determination of creatinine levels in serum and urine for the diagnosis of renal dysfunction. CSHase was discovered in Arthrobacter sp., pseudomonas putida 77 and other microorganisms. Kim et al. (1986) purified and characterized CSHase from P. putida 77. The CSHase from Arthrobacter sp. was also characterized and finally expressed in Escherichia coli.

Besides CSHase there are a number of N-carbamoylamide amino-hydrolase known, such as b -ureidopropionase (EC 3.5.1.6), ureido-succinase (EC 3.5.1.7), citrullinase (EC 3.5.1.20) and other N-carbamoyl amino acid amidohydrolase with D and L specicities from different microorganisms. All these enzymes possess a common N-carbamoylamide-hydrolyzing function but differ from CSHase in specificity and other properties. b-Ureidopropionase, which hydrolyzes N-carbamoyl-b-alanine, is much lager in molecular mass and, in contrast to CSHase, is not affected by sulfhydryl reagents. Ureidosuccinase requires divalent metal ions for its reaction with N-carbamoylaspartic acid, which is not the case for CSHase. Citrullinase decarbamoylates L-citrulline to form ornithine.
The N-carbamoyl-D-amino acid amidohydrolase from Comamonas sp. characterized by Ogawa et al. (1993), hydrolyzes a variety of N-carbamoyl-D-amino acids, show a molecular mass close to that of CSHase, does not require metal ions for activity, is sensitive to thiol reagents like CSHase, but is not able to hydrolyze N-carbamoylsarcosine. The NH2-terminal sequence(30 residues) shows no similarity to CSHase and other amidohydrolase. So far no apparent homology in mechanism or structure can be found among all these decarbamylases.
The X-ray structure of bacterial creatinase (amidinohydrolase) hase been solved and, on the basis of the structures of several inhibitor and substrate complexes, it was possible to propose an enzymatic mechanism explaning the hydrolysis of the guanidinium moiety of creatine: the resonance-stabilized guanidinium group is attacked by a histidine-activated water molecule as nucleophile.
The crystal structure of N-carbamoylsarcosine amidohydrolase (CSHase) from Arthrobacter sp. Was determined in order to analyze its functions and possible mechanistic similarities to the known amidohydrolase. Here are two new crystal structures of CSHase in complex with active-site-directed irreversible inhibitors, which can be identified the active site of CSHase.
¡@
¡@
¡@
CSHase is strongly inhibited by glyoxylate in the competivite test assay. The enzyme contains two cysteine residues per subunit (Cys65 and Cys177),which can easily be determined using 5,5¢ -dithiobis(2-nitrobenzoic acid)(DTNB) as sulfhydryl reagent under denaturing conditions (at least 2 M urea), where all cysteine residues are accessible. In the native state, however, only half of the sulfhydryl content could be determined with DTNB. In the presence of a large excess of substrate, no free cysteine could be detected during the steady-state phase of the enzymatic reaction, indicating that Cys177 is involved in catalysis.
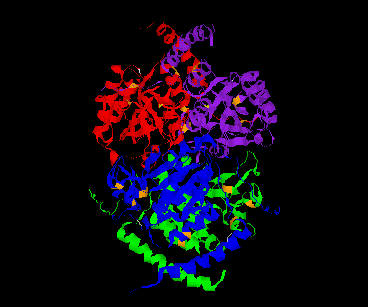
Dimer A, B Dimer C,D
¡@
¡@
The two active-site inhibitors of the enzyme(succinic semialdehyde and glyoxylate) are both aldehydes which inactivate the enzyme irreversibly through covalent binding to the cysteine residues Cys177.
The inhibitors were added by soaking the CSHase crystals, but the weak reversible inhibitors did not bind under these conditions. Cocrystallization of the protein with those weak ligands also showed no binding.
¡@
¡@
¡@
The CSHase crystal structure model consists of four subunits, 431 water molecules and four sulfate ions bound to the catalytic site of each subunit. The atoms of the model are well defined except for seven N-terminal and five C-terminal amino acid residues, which are disordered. The monomer has an a /b type structure, consisting of a central six-stranded parallel b -pleated sheet with a strong left-handed twist. Three of them on one side are involved in non-covalent, tight intersubunit binding (A-B and C-D), whereas two helices on the other side of the b -sheet are exposed to the solvent. Only a few intersubunit contacts occur between the compact dimer AB and CD, so that the whole tetramer appears as a dimer of dimers. The folding of a dimer and the tetramer of CSHase as Ca -trace and ribbon drawings are shown in Figures 2 and 3, respectively.
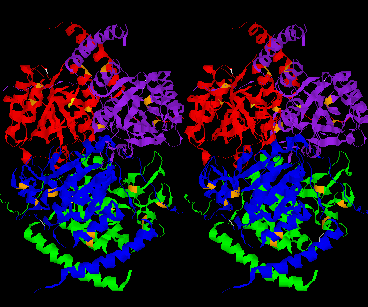
The four active sites of CSHase are situated at the interface of the compact dimers AB and CD. Within a monomer, the sulfate and substrate binding site is located at the carboxy end of a b -strand (residues 166 to 172), where the strand changes direction and is connected to a helix (residues 177 to 188). At the end of the b -strand, there is a cis-peptide bond between the residues Ala172 and Thr173, which form the " bottom" of the catalytic site. The helix is involved in intersubunit contacts of the compact dimers AB and CD and carries the active-site cysteine residue(Cys177). Mercury ions bind to the thiol group of that active-site cysteine (Cys177) at subunit A and D, but not to subunits B and C. All four active sites in the tetramer of CSHase contain a sulfate ion and a water molecule. The sulfate is hydrogen-bounded to the amidino group of residue Arg202, to a lysine residue from the neighboring subunit (Lys217), and to Ne of residue Trp56.Further bonds are to the main-chain nitrogen atoms of residue Thr173 (the cis-peptide bond) and the active-site cysteine residue Cys177. The entrance to the active site guarded by residue Arg202, which must move to allow diffusion of ligands into the catalytic cleft.
¡@
¡@
¡@
Crystal structures of N-carbamoylsarcosine amidohydrolase (CSHase; EC 3.5.59) have been analyzed by C -ray diffraction methods with two different inhibitors bound to the active site at 2.28 Å and 2.37 Å resolution. The catalytic center of the enzyme could be identified on the basis of these structures. The four substrate binding sites are situated at the intersubunit interfaces of the compact dimer AB and CD of the homotetrameric enzyme. Both inhibitors inactivate the enzyme irreversibly through covalent binding of their aldehyde groups to the thiol group of the active -site cysteine residue Cys177. Within the identified substrate binding sites a number of residues from different subunits are involved in hydrogen bonding of the inhibitors. Two residues (Ala172 and Thr173) that form an unusual cis-peptide bond at the binding site are important components in fixing the examined inhibitors by hydrogen bonds. An electrochemical enzyme assay for CSHase was used to test the effect of inhibitors and substrate analogs on the enzyme¢ s activity, revealing the high substrate specificity of CSHase. The intrinsic tryptophan fluorescence of CSHase increases strongly upon substrate and inhibitor binding. As most of the tryptophyl residues are located at the active sites, they are thus considerably affected by ligand binding. Fluorescence-detected stopped-flow measurements have been used to study the kinetics of glyoxylate and substrate binding to CSHase. Substrate and inhibitor binding could clearly be distinguished in the stopped-flow experiments. Inhibitor binding reveals at least three different elementary processes, whereas substrate binding is much faster and contains phases with different sign in amplitude.
¡@
¡@
Enzyme: 3.5.1.59: N-CARBAMOYLSARCOSINE AMIDOHYDROLASE.
¡@
¡@
¡@
Zajc, A., Romão, M. J., Turk, B. & Huber, R.(1996). Crystallographic and Fluorescence Studies of Ligand Binding to N-Carbamoylsarcosine Amidohydrolase from Arthrobacter sp.. J. Mol. Biol. 263, 269-283.
Romão, M. J., Turk, D., Gomis-Rüth, F. X., Huber, R.,Schumacher, G., Möllering, H. & Rüssmann, L.(1992). Crystal structure analysis, refinement and enzymatic reaction mechanism of N-carbamoylsarcosine amidohydrolase from Arthrobacter sp. at 2.0Å resolution. J. Mol. Biol. 226, 1111-1130.
¡@
¡@
¡@
¡@
¡@
¡@
¡@