
**********************************
* EF-hand calcium-binding domain *
**********************************
Many calcium-binding proteins belong to the same evolutionary family and share
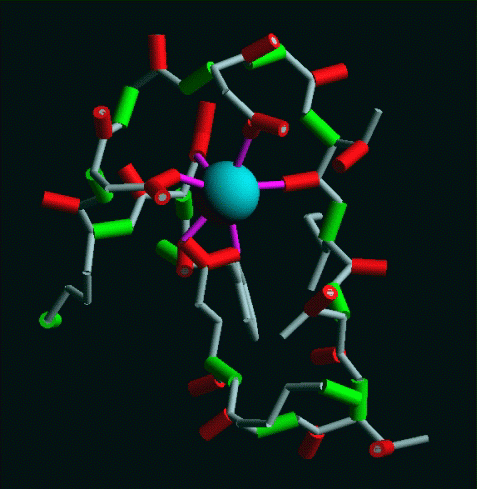
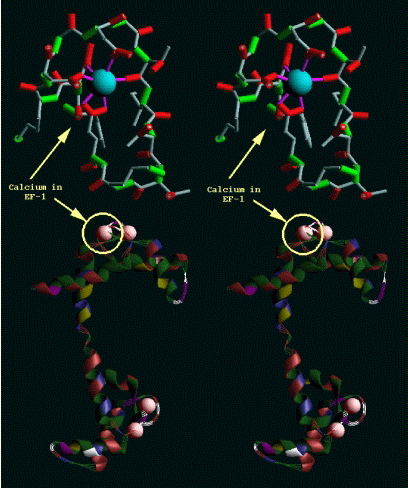
a type of calcium-binding domain known as the EF-hand [1 to 5]. This type of
domain consists of a twelve residue loop flanked on both side by a twelve
residue alpha-helical domain. In an EF-hand loop the calcium ion is
octahedrally coordinated. The six residues involved in the binding are in
positions 1, 3, 5, 7, 9 and 12; these residues are denoted by X, Y, Z, -Y, -X
and -Z.
We list below the proteins which are known to contain EF-hand regions. For
each type of protein we have indicated between parenthesis the total number of
EF-hand regions known or supposed to exist. This number does not include
regions which clearly have lost their calcium-binding properties, or the
atypical low-affinity site (which spans thirteen residues) found in the S-100/
ICaBP family of proteins [6].
- Aequorin and Renilla luciferin binding protein (LBP) (Ca=3).
- Alpha actinin (Ca=2).
- Calbindin (Ca=4).
- Calcineurin B subunit (protein phosphatase 2B regulatory subunit) (Ca=4).
- Calcium-binding protein from Streptomyces erythraeus (Ca=3?).
- Calcium-binding protein from Schistosoma mansoni (Ca=2?).
- Calcium-binding proteins TCBP-23 and TCBP-25 from Tetrahymena thermophila
(Ca=4?).
- Calcium-dependent protein kinases (CDPK) from plants (Ca=4).
- Calcium vector protein from amphoxius (Ca=2).
- Calcyphosin (thyroid protein p24) (Ca=4?).
- Calmodulin (Ca=4, except in yeast where Ca=3).
- Calpain small and large chains (Ca=2).
- Calretinin (Ca=6).
- Calcyclin (prolactin receptor associated protein) (Ca=1).
- Caltractin (centrin) (Ca=2 or 4).
- Cell Division Control protein 31 (gene CDC31) from yeast (Ca=2?).
- Diacylglycerol kinase (EC 2.7.1.107) (DGK) (Ca=2).
- FAD-dependent glycerol-3-phosphate dehydrogenase (EC 1.1.99.5) from
mammals (Ca=1).
- Fimbrin (plastin) (Ca=2).
- Flagellar calcium-binding protein (1f8) from Trypanosoma cruzi (Ca=1 or 2).
- Guanylate cyclase activating protein (GCAP) (Ca=3).
- Intestinal calcium-binding protein (ICaBPs) (Ca=1).
- MIF related proteins 8 (MRP-8 or CFAG) and 14 (MRP-14) (Ca=1).
- Myosin regulatory light chains (Ca=1).
- Oncomodulin (Ca=2).
- Osteonectin (basement membrane protein BM-40) (SPARC) and proteins that
contains an 'osteonectin' domain (QR1, matrix glycoprotein SC1) (see the
entry <PDOC00535>) (Ca=1).
- Parvalbumins alpha and beta (Ca=2).
- Placental calcium-binding protein (18a2) (nerve growth factor induced
protein 42a) (p9k) (Ca=1).
- Recoverins (visinin, hippocalcin, neurocalcin, S-modulin) (Ca=2 to 3).
- Reticulocalbin (Ca=4).
- S-100 protein, alpha and beta chains (Ca=1).
- Sarcoplasmic calcium-binding protein (SCPs) (Ca=2 to 3).
- Sea urchin proteins Spec 1 (Ca=4), Spec 2 (Ca=4?), Lps-1 (Ca=8).
- Serine/threonine protein phosphatase rdgc (EC 3.1.3.16) from Drosophila
(Ca=2)
- Sorcin V19 from hamster (Ca=2).
- Spectrin alpha chain (Ca=2).
- Squidulin (optic lobe calcium-binding protein) from squid (Ca=4).
- Troponins C; from skeletal muscle (Ca=4), from cardiac muscle (Ca=3), from
arthropods and molluscs (Ca=2).
There has been a number of attempts [7,8] to develop patterns that pick-up EF-
hand regions, but these studies were made a few years ago when not so many
different families of calcium-binding proteins were known. We therefore
developed a new pattern which takes into account all published sequences. This
pattern includes the complete EF-hand loop as well as the first residue which
follows the loop and which seem to always be hydrophobic.
-Consensus pattern:
1 2 3 4 5 6 7 8 9 10 12
X Y Z -Y -X -Z
D-x-[DNS]-{ILVFYW}-[DENSTG]-[DNQGHRK]-{GP}-[LIVMC]-[DENQSTAGC]-x(2)-[DE]-
13
[LIVMFYW]
-Sequences known to belong to this class detected by the pattern: ALL, except
for chicken actinin and human S-100P.
-Other sequence(s) detected in SWISS-PROT: 52 proteins which are probably not
calcium-binding and a few proteins for which we have reason to believe that
they bind calcium: a number of endoglucanases and a xylanase from the
cellulosome complex of Clostridium [9] and two different isozymes of inositol
phospholipid-specific phospholipase C [10].
-Note: positions 1 (X), 3 (Y) and 12 (-Z) are the most conserved.
-Note: the 6th residue in an EF-hand loop is, in most cases a Gly, but the
number of exceptions to this 'rule' has gradually increased and we felt that
the pattern should include all the different residues which have been shown
to exist in this position in functional Ca-binding sites.
-Note: the pattern will, in some cases, miss one of the EF-hand regions in
some proteins with multiple EF-hand domains.
-Expert(s) to contact by email: Cox J.A.
cox@sc2a.unige.ch
Kretsinger R.H.
rhk5i@virginia.edu
-Last update: November 1995 / Text revised.
[ 1] Kawasaki H., Kretsinger R.H.
Protein Prof. 2:305-490(1995).
[ 2] Kretsinger R.H.
Cold Spring Harbor Symp. Quant. Biol. 52:499-510(1987).
[ 3] Moncrief N.D., Kretsinger R.H., Goodman M.
J. Mol. Evol. 30:522-562(1990).
[ 4] Nakayama S., Moncrief N.D., Kretsinger R.H.
J. Mol. Evol. 34:416-448(1992).
[ 5] Heizmann C.W., Hunziker W.
Trends Biochem. Sci. 16:98-103(1991).
[ 6] Kligman D., Hilt D.C.
Trends Biochem. Sci. 13:437-443(1988).
[ 7] Gariepy J., Hodges R.S.
FEBS Lett. 160:1-6(1983).
[ 8] Haiech J., Sallantin J.
Biochimie 67:555-560(1985).
[ 9] Chauvaux S., Beguin P., Aubert J.-P., Bhat K.M., Gow L.A., Wood T.M.,
Bairoch A.
Biochem. J. 265:261-265(1990).
[10] Bairoch A., Cox J.A.
FEBS Lett. 269:454-456(1990).