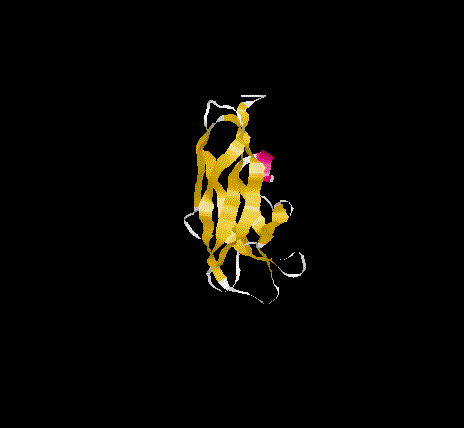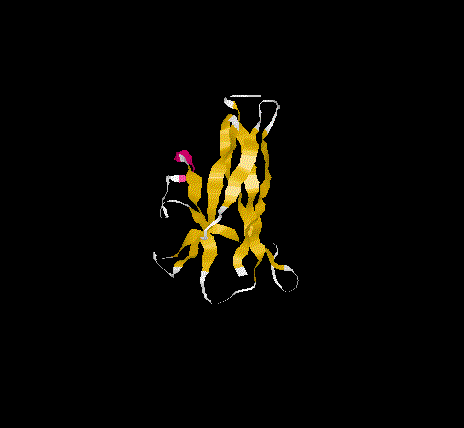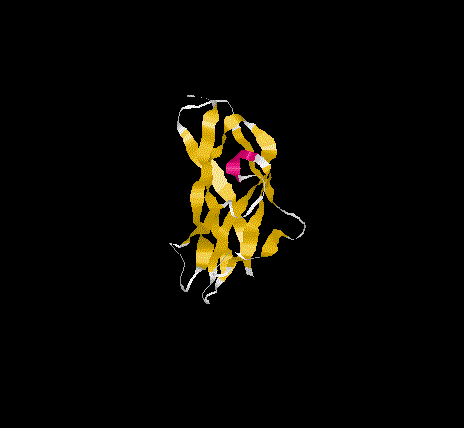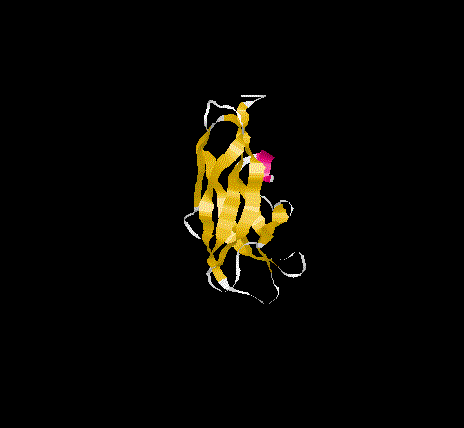
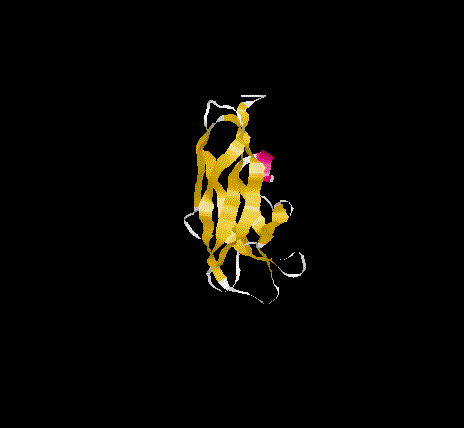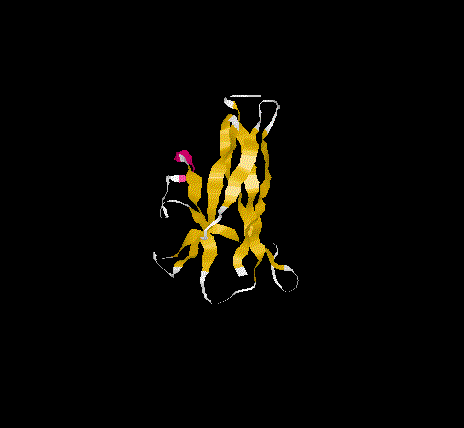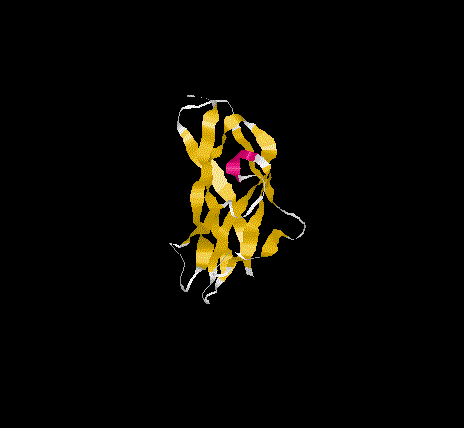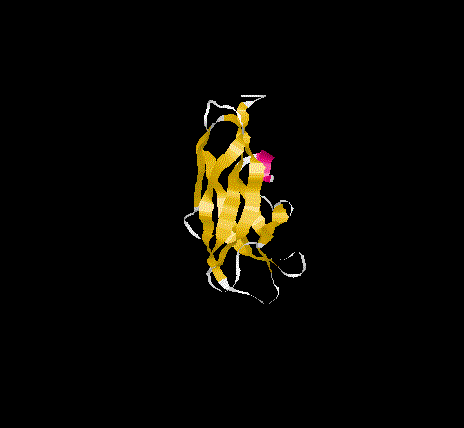
¡@
Starch Binding Domain
¡@
The overall topology of the SBD of G1 from A. niger shows eight b-
strands forming two major b-sheets ( Jacks et al., 1995). One sheet
consists of five strands(2, 3, 5, 6, and 7) arranged in antiparallel
fashion while the second sheet(1, 4, and 8)has one parallel and one
antiparallel strand pair. Residues A523, P561 and L562 represent
additional b-sheet residues to those previously identified
( Jacks et al., 1995) due to an extension of the antiparallel
interaction of strand 1 and 4. The b-strands are well stabilised as
evidenced by the slow amide exchange rates for these residues, which
in some cases have half-lives of weeks or months at 313k and pH 5.2,
and form the core of the SBD. The longer strands(1,4 ,5 and 8) show
a degree of curvature and twist which allows better packing.
In addition, two hydrogen bonds between strand 3 and 4 involve residues
S552 and Y564 in antiparallel fashion(Sorimachi et al., 1996).Thus the
SBD forms an open-sided, distorted, b-barrel structure.
¡@
Possible binding site residues
The substrate binding sites in domain E of CGTase(cyclodextrin
glycosyltransferase, EC2.4.1.19) is homologous to the SBD of glucoamylase
from A. niger showing aprroximately 37% amino acids sequence identity,and
it have been studied previously in detail by Lawson et al. (1994) in the
crystal structure of the CGTase/maltose complex.Thus we can use the
knowlege of the SBD disulfide bridge, CGTase binding site residues and
homology between SBD and CGTase residues amongst other criteria to predict
the possible binding site residues. The proposed SBD residues were for site 1,
W543, E576, K578, W590, N595 and for site 2, T525, T526, Y527, G528, E529,
N530, D554, K555, D560, W563.
¡@
¡@
¡@