2. Transcription of the ant gene during lytic growth of bacteriophage P22 is regulated by the cooperative binding of two Arc repressor dimers to a 21-base-pair operator site. Please find the co-crystal structure of this Arc tetramer-operator complex from PDB.
(a) Make an animation (by Gifbuilder) of whole complex.
(b) Show the DNA sequences of the Arc operator.
- Chain E: Nucleic acid sequence ( 22 bases )
- Sequence:
T-A-T-A-G-T-A-G-A-G-T-G-C-T-T-C-T-A-T
-C-A-T
- Sequence:
T-A-T-A-G-T-A-G-A-G-T-G-C-T-T-C-T-A-T
-C-A-T
-
Chain F: Nucleic acid sequence ( 22 bases )
- Sequence: A-A-T-G-A-T-A-G-A-A-G-C-A-C-T-C-T-A-C -T-A-T
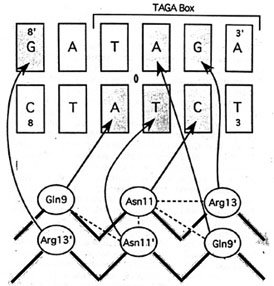
(c) The picture below is the hydrogen bonds between beta-sheet side chains and DNA bases. Show three side chains (gln9, asn11, arg13) from one monomer and corresponding DNA bases of them on your home page. Color three side chain with different color. Measure the distance between possible hydrogen bonds.
![]() Gln 9D - A7F
Gln 9D - A7F ![]() Asn 11D - T17E
Asn 11D - T17E ![]() Arg 13D - G4F
Arg 13D - G4F