Multiple Sequence Alignment of MLH1
protein.
- To do Multiple Sequence Alignment in Biology Workbench
- To draw phylogenetic tree from alignment
¡@
1. Add MLH1_Human protein to the Biology Workbench. Predict its secondary structure by GOR4.
>MLH1_HUMAN
Sequence MSFVAGVIRR LDETVVNRIA AGEVIQRPAN AIKEMIENCL DAKSTSIQVI
Structure CCCCEEEEEE CCHHHHHHHH HCHHHHHHHH HHHHHHHHHC CCCCCCHHHH
Sequence VKEGGLKLIQ IQDNGTGIRK EDLDIVCERF TTSKLQSFED LASISTYGFR
Structure HHHCCCEEEE ECCCCCCCHH HHHHHHHCCC CCCCCCCCHH HHHHCCCCCC
Sequence GEALASISHV AHVTITTKTA DGKCAYRASY SDGKLKAPPK PCAGNQGTQI
Structure CCHHHHHHHE EEEEEEECCC CCCCEEEECC CCCCCCCCCC CCCCCCCCEE
Sequence TVEDLFYNIA TRRKALKNPS EEYGKILEVV GRYSVHNAGI SFSVKKQGET
Structure EEHHHHHHHH HHHHHHCCCC HHHHHEEEEE ECCCCCCCCE EEEECCCCCE
Sequence VADVRTLPNA STVDNIRSIF GNAVSRELIE IGCEDKTLAF KMNGYISNAN
Structure EEEEEECCCC CCCCCEEEEC CCCCCHHHHH HCCCHHHHHH CCCCCEECCC
Sequence YSVKKCIFLL FINHRLVEST SLRKAIETVY AAYLPKNTHP FLYLSLEISP
Structure CCCCCEEEEE ECCCCHHHHH HHHHHHHHHH HHCCCCCCCC EEEECCCCCC
Sequence QNVDVNVHPT KHEVHFLHEE SILERVQQHI ESKLLGSNSS RMYFTQTLLP
Structure CCCCEEECCC CCHHHHHHHH HHHHHHHHHH HHHHHCCCCC CEEEEEEECC
Sequence GLAGPSGEMV KSTTSLTSSS TSGSSDKVYA HQMVRTDSRE QKLDAFLQPL
Structure CCCCCCCCEE EEEEEEEEEC CCCCCCHHHH HHHHHHHHHH HHHHHHHCCC
Sequence SKPLSSQPQA IVTEDKTDIS SGRARQQDEE MLELPAPAEV AAKNQSLEGD
Structure CCCCCCCCCE EECCCCCCHH HHHHHHHHHH HHHCCCHHHH HHHHHCCCCC
Sequence TTKGTSEMSE KRGPTSSNPR KRHREDSDVE MVEDDSRKEM TAACTPRRRI
Structure CCCCCCHHHC CCCCCCCCCC CCCCCCCCHH HHHHHHHHHH HHHCCCCCEE
Sequence INLTSVLSLQ EEINEQGHEV LREMLHNHSF VGCVNPQWAL AQHQTKLYLL
Structure ECCCCHHHHH HHHHHHHHHH HHHHHCCCCE EEEECCCCHH HHHHHHHHHH
Sequence NTTKLSEELF YQILIYDFAN FGVLRLSEPA PLFDLAMLAL DSPESGWTEE
Structure HCCCCHHHHH HHHHHHCCCC CCEECCCCCC CHHHHHHHHC CCCCCCCCCC
Sequence DGPKEGLAEY IVEFLKKKAE MLADYFSLEI DEEGNLIGLP LLIDNYVPPL
Structure CCCCCCHHHH HHHHHHHHHH HHHHHHHHHH HHCCCCCCCC EEECCCCCCC
Sequence EGLPIFILRL ATEVNWDEEK ECFESLSKEC AMFYSIRKQY ISEESTLSGQ
Structure CCCCHHHHHH HHHHCHHHHH HCCCCCCCCC HHHHHCCCCC CCHHHHCCCC
Sequence QSEVPGSIPN SWKWTVEHIV YKALRSHILP PKHFTEDGNI LQLANLPDLY
Structure CCCCCCCCCC CCCEEEECHH HHHHHCCCCC CCCCCCCCHH HHHHCCCCCE
Sequence KVFERC
Structure EEEEEC
LEGEND:
Alpha Helix = H Beta Sheet = E Random Coil = C
¡@
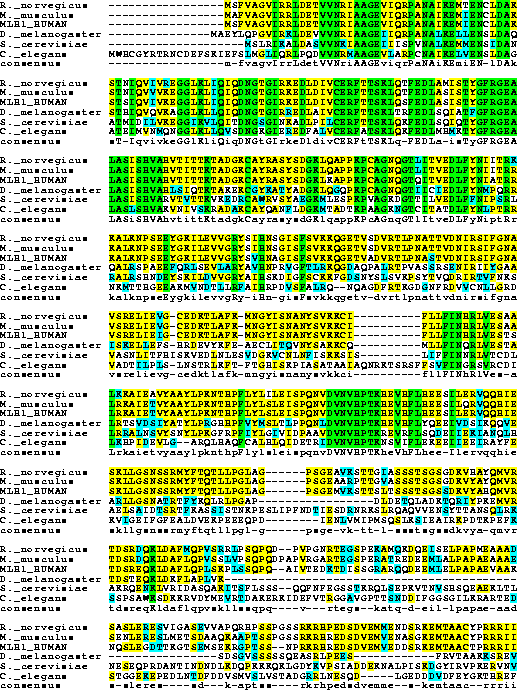
2. Do a homology searching of MLH1_Human in Genpept Full Release
Database. Import MLH1-like protein of C. elegans, S. cerevisiae, D. melanogaster,
R. norvegicus and M. musculus to your workbench. Run CLUSTALW to get multiple
sequence alignment for these six proteins.
¡@
Consensus key (see documentation for details)
* - single, fully conserved residue
: - conservation of strong groups
. - conservation of weak groups
- no consensus
CLUSTAL W (1.81) multiple sequence alignment
GENPEPT_460627 --------------------MSLRIKALDASVVNKIAAGEIIISPVNALKEMMENSIDAN
GENPEPT_825572 --------------------MSLRIKALDASVVNKIAAGEIIISPVNALKEMMENSIDAN
GENPEPT_7595954 -----------------MAFVAGVIRRLDETVVNRIAAGEVIQRPANAIKEMIENCLDAK
GENPEPT_1724118 -----------------MSFVAGVIRRLDETVVNRIAAGEVIQRPANAIKEMTENCLDAK
MLH1_HUMAN -----------------MSFVAGVIRRLDETVVNRIAAGEVIQRPANAIKEMIENCLDAK
GENPEPT_3192877 ---------------MAEYLQPGVIRKLDEVVVNRIAAGEIIQRPANALKELLENSLDAQ
GENPEPT_3880333 MWHCGYRTRNCDEFSKIEFSLMGLIQRLPQDVVNRMAAGEVLARPCNAIKELVENSLDAG
*: * ***::****:: * **:**: **.:**
GENPEPT_460627 ATMIDILVKEGGIKVLQITDNGSGINKADLPILCERFTTSKLQKFEDLSQIQTYGFRGEA
GENPEPT_825572 ATMIDILVKEGGIKVLQITDNGSGINKADLPILCERFTTSKLQKFEDLSQIQTYGFRGEA
GENPEPT_7595954 STNIQVVVKEGGLKLIQIQDNGTGIRKEDLDIVCERFTTSKLQTFEDLASISTYGFRGEA
GENPEPT_1724118 STNIQVIVREGGLKLIQIQDNGTGIRKEDLDIVCERFTTSKLQTFEDLAMISTYGFRGEA
MLH1_HUMAN STSIQVIVKEGGLKLIQIQDNGTGIRKEDLDIVCERFTTSKLQSFEDLASISTYGFRGEA
GENPEPT_3192877 STHIQVQVKAGGLKLLQIQDNGTGIRREDLAIVCERFTTSKLTRFEDLSQIATFGFRGEA
GENPEPT_3880333 ATEIMVNMQNGGLKLLQVSDNGKGIEREDFALVCERFATSKLQKFEDLMHMKTYGFRGEA
:* * : :: **:*::*: ***.**.: *: ::****:**** **** : *:******
GENPEPT_460627 LASISHVARVTVTTKVKEDRCAWRVSYAEGKMLESPKPVAGKDGTTILVEDLFFNIPSRL
GENPEPT_825572 LASISHVARVTVTTKVKEDRCAWRVSYAEGKMLESPKPVAGKDGTTILVEDLFFNIPSRL
GENPEPT_7595954 LASISHVAHVTITTKTADGKCAYRASYSDGKLQAPPKPCAGNQGTLITVEDLFYNIITRR
GENPEPT_1724118 LASISHVAHVTITTKTADGKCAYRASYSDGKLQAPPKPCAGNQGTLITVEDLFYNIITRK
MLH1_HUMAN LASISHVAHVTITTKTADGKCAYRASYSDGKLKAPPKPCAGNQGTQITVEDLFYNIATRR
GENPEPT_3192877 LASISHVAHLSIQTKTAKEKCGYKATYADGKLQGQPKPCAGNQGTIICIEDLFYNMPQRR
GENPEPT_3880333 LASLSHVAKVNIVSKRADAKCAYQANFLDGKMTADTKPAAGKNGTCITATDLFYNLPTRR
***:****::.: :* . :*.::..: :**: .** **::** * ***:*: *
GENPEPT_460627 RALRSHNDEYSKILDVVGRYAIHSKDIGFSCKKFGDSNYSLSVKPSYTVQDRIRTVFNKS
GENPEPT_825572 RALRSHNDEYSKILDVVGRYAIHSKDIGFSCKKFGDSNYSLSVKPSYTVQDRIRTVFNKS
GENPEPT_7595954 KALKNPSEEYGKILEVVGRYSIHNSGISFSVKKQGETVSDVRTLPNATTVDNIRSIFGNA
GENPEPT_1724118 KALKNPSEEYGKILEVVGRYSIHNSGISFSVKKQGETVSDVRTLPNATTVDNIRSIFGNA
MLH1_HUMAN KALKNPSEEYGKILEVVGRYSVHNAGISFSVKKQGETVADVRTLPNASTVDNIRSIFGNA
GENPEPT_3192877 QALRSPAEEFQRLSEVLARYAVHNPRVGFTLRKQGDAQPALRTPVASSRSENIRIIYGAA
GENPEPT_3880333 NKMTTHGEEAKMVNDTLLRFAIHRPDVSFALRQ--NQAGDFRTKGDGNFRDVVCNLLGRD
. : . :* : :.: *:::* :.*: :: : . . . : : : .
GENPEPT_460627 VASNLITFHISKVEDLNLESVDGKVCNLNFISKKSIS---------LIFFINNRLVTCDL
GENPEPT_825572 VASNLITFHISKVEDLNLESVDGKVCNLNFISKKSIS---------PIFFINNRLVTCDL
GENPEPT_7595954 VSRELIEVG-CEDKTLAFK-MNGYISNANYSVKKCI----------FLLFINHRLVESAA
GENPEPT_1724118 VSRELIEVG-CEDKTLAFK-MNGYISNANYSVKKCI----------FLLFINHRLVESAA
MLH1_HUMAN VSRELIEIG-CEDKTLAFK-MNGYISNANYSVKKCI----------FLLFINHRLVESTS
GENPEPT_3192877 ISKELLEFS-HRDEVYKFE-AECLITQVNYSAKKCQ----------MLLFINQRLVESTA
GENPEPT_3880333 VADTILPLS-LNSTRLKFT-FTGHISKPIASATAAIAQNRKTSRSFFSVFINGRSVRCDI
:: :: . . : : : . . .*** * * .
GENPEPT_460627 LRRALNSVYSNYLPKGFRPFIYLGIVIDPAAVDVNVHPTKREVRFLSQDEIIEKIANQLH
GENPEPT_825572 LRRALNSVYSNYLPKGNRPFIYLGIVIDPAAVDVNVHPTKREVRFLSQDEIIEKIANQLH
GENPEPT_7595954 LRKAIETVYAAYLPKNTHPFLYLSLEISPQNVDVNVHPTKHEVHFLHEESILQRVQQHIE
GENPEPT_1724118 LKKAIEAVYAAYLPKNTHPFLYLILEISPQNVDVNVHPTKHEVHFLHEESILERVQQHIE
MLH1_HUMAN LRKAIETVYAAYLPKNTHPFLYLSLEISPQNVDVNVHPTKHEVHFLHEESILERVQQHIE
GENPEPT_3192877 LRTSVDSIYATYLPRGHHPFVYMSLTLPPQNLDVNVHPTKHEVHFLYQEEIVDSIKQQVE
GENPEPT_3880333 LKHPIDEVLG--ARQLHAQFCALHLQIDETRIDVNVHPTKNSVIFLEKEEIIEEIRAYFE
*: .:: : . : * : : : :********..* ** ::.*:: : ..
GENPEPT_460627 AELSAIDTSRTFKASSISTNKPESLIPFNDTIESDRNRKSLRQAQVVENSYTTANSQLRK
GENPEPT_825572 AELSAIDTSRTFKASSISTNKPESLIPFNDTIESDRNRKSLRQAQVVENSYTTANSQLRK
GENPEPT_7595954 SKLLGSNSSRMYFTQTLLPGLAG------PSGEAARPTTGVASSSTSGSGDKVYAYQMVR
GENPEPT_1724118 SKLLGSNSSRMYFTQTLLPGLAG------PSGEAVKSTTGIASSSTSGSGDKVHAYQMVR
MLH1_HUMAN SKLLGSNSSRMYFTQTLLPGLAG------PSGEMVKSTTSLTSSSTSGSSDKVYAHQMVR
GENPEPT_3192877 ARLLGSNATRTFYKQLRLPGAP-----------------DLDETQLADKTQRIYPKEMVR
GENPEPT_3880333 KVIGEIFGFEALDVEKPEEEQPD--------IENLVMIPMSQSLKSIEAIRKPDTKPEFK
: . . . . . :
GENPEPT_460627 AKRQENKLVRIDASQAKITSFLSSS--QQFNFEGSSTKRQLSEPKVTNVSHSQEAEKLTL
GENPEPT_825572 AKRQENKLVRIDASQAKITSFLSSS--QQFNFEGSSTKRQLSEPKVTNVSHSQEAEKLTL
GENPEPT_7595954 TDSRDQKLDAFLQPVSSLVPSQPQDPAPVRGARTEGSPERATREDEEMLALPAPAEAAAE
GENPEPT_1724118 TDSRDQKLDAFMQPVSRRLPSQPQD--PVPGNRTEGSPEKAMQKDQEISELPAPMEAAAD
MLH1_HUMAN TDSREQKLDAFLQPLSKPLSSQPQ--AIVTEDKTDISSGRARQQDEEMLELPAPAEVAAK
GENPEPT_3192877 TDSTEQKLDKFLAPLVK-------------------------------------------
GENPEPT_3880333 SSPSAWKSDKKRVDYMEVRTDAKERKIDEFVTRGGAVGPTTSNDDIFGGSGILKRARTED
:. *
GENPEPT_460627 NESEQPRDANTINDNDLKDQPKKKQKLGDYKVPSIADDEKNALPISKDGYIRVPKERVNV
GENPEPT_825572 NESEQPRDANTINDNDLKDQPKKKQKLGDYKVPSIADDEKNALPISKDGYIRVPKERVNV
GENPEPT_7595954 SENLERESLMETSDAAQKAAPTSSPGSSRKRHREDSDVEMVENASGKEMTAACYPRRRII
GENPEPT_1724118 SASLERESVIGASEVVAPQRHPSSPGSSRKRHPEDSDVEMMENDSRKEMTAACYPRRRII
MLH1_HUMAN NQSLEGDTTKGTSEMSEKRGPTSS--NPRKRHREDSDVEMVEDDSRKEMTAACTPRRRII
GENPEPT_3192877 ----------------SDSGVSSSSSQEASRLPEES------------FRVTAAKKSREV
GENPEPT_3880333 STGGEKEPEDLNTDFDDVSMVSLVSTADGRRLNES-----QDLGEDDDVDFEYGKTHREF
: . .
GENPEPT_460627 NLTSIKKLREKVDDSIHRELTDIFANLNYVGVVDEERRLAAIQHDLKLFLIDYGSVCYEL
GENPEPT_825572 NLTSIKKLREKVDDSIHRELTDIFANLNYVGVVDEERRLAAIQHDLKLFLIDYGSVCYEL
GENPEPT_7595954 NLTSVLSLQEEISERCHETLREILRNHSFVGCVNPQW--ALAQHQTKLYLLNTTKLSEEL
GENPEPT_1724118 NLTSVLSLQEEINDRGHETLREMLRNHTFVGCVNPQW--ALAQHQTKLYLLNTTKLSEEL
MLH1_HUMAN NLTSVLSLQEEINEQGHEVLREMLHNHSFVGCVNPQW--ALAQHQTKLYLLNTTKLSEEL
GENPEPT_3192877 RLSSVLDMRKRVERQCSVQLRSTLKNLVYVGCVDERR--ALFQHETRLYMCNTRSFSEEL
GENPEPT_3880333 HFESIEVLRKEIIANSSQSLREMFKTSTFVGSINVKQ--VLIQFGTSLYHLDFSTVLREF
.: *: :::.: * . : . :** :: . . *. *: : .. *:
GENPEPT_460627 FYQIGLTDFANFGKINLQSTNVSDDIVLYNLLSEFDELN-DDASK---------EKIISK
GENPEPT_825572 FYQIGLTDFANFGKINLQSTNVSDDIVLYNLLSEFDELN-DDASK---------EKIISK
GENPEPT_7595954 FYQILIYDFANFGVLRLSEPAPLFDLAMLALDSPESGWTEDDGPKEGLA-----EYIVEF
GENPEPT_1724118 FYQILIYDFANFGVLRLPEPAPLFDFAMLALDSPESGWTEEDGPKEGLA-----EYIVEF
MLH1_HUMAN FYQILIYDFANFGVLRLSEPAPLFDLAMLALDSPESGWTEEDGPKEGLA-----EYIVEF
GENPEPT_3192877 FYQRMIYEFQNCSEITICPPLPLKELLILSLESRAAGWTPEDEDKAELA-----DGAADI
GENPEPT_3880333 FYQISVFSFGNYGSYRLDE-EPPAIIEILELLGELSTREPNYAAFEVFANVENRFAAEKL
*** : .* * . : : : * . : .
GENPEPT_460627 IWDMSSMLNEYYSIELVNDGLDNDLKSVKLKSLPLLLKGYIPSLVKLPFFIYRLGKEVDW
GENPEPT_825572 IWDMSSMLNEYYSIELVNDGLDNDLKSVKLKSLPLLLKGYIPSLVKLPFFIYRLGKEVDW
GENPEPT_7595954 LKKKAEMLADYFSVEIDEEGN--------LIGLPLLIDSYVPPLEGLPIFILRLATEVNW
GENPEPT_1724118 LKKKAKMLADYFSVEIDEEGN--------LIGLPLLIDSYVPPLEGLPIFILRLATEVNW
MLH1_HUMAN LKKKAEMLADYFSLEIDEEGN--------LIGLPLLIDNYVPPLEGLPIFILRLATEVNW
GENPEPT_3192877 LLKKAPIMREYFGLRISEDGM--------LESLPSLLHQHRPCVAHLPVYLLRLATEVDW
GENPEPT_3880333 LAEHADLLHDYFAIKLDQLENGR----LHITEIPSLVHYFVPQLEKLPFLIATLVLNVDY
: . : :: :*:.:.: : : :* *:. . * : **. : * :*::
GENPEPT_460627 EDEQECLDGILREIALLYIPDMVPKVDTLDASLSEDEKAQFINRKEHISSLLEHVLFPCI
GENPEPT_825572 EDEQECLDGILREIALLYIPDMVPKVDTSDASLSEDEKAQFINRKEHISSLLEHVLFPCI
GENPEPT_7595954 DEEKECFESLSKECAMFYSIRKQYILEESTLSGQQSDMPGSTSKPWKWT--VEHIIYKAF
GENPEPT_1724118 DEE-ECFESLSKECAVFYSIRKQYILEESALSGQQSDMPGSPSKPWKWT--VEHIIYKAF
MLH1_HUMAN DEEKECFESLSKECAMFYSIRKQYISEESTLSGQQSEVPGSIPNSWKWT--VEHIVYKAL
GENPEPT_3192877 EQETRCFETFCRETARFY--------------AQLDWREGATAVFSRWT--MEHVLFPAF
GENPEPT_3880333 DDEQNTFRTICRAIGDLFTLDTN---------FITLDKKISAFSATPWKTLIKEVLMPLV
::* . : : : . :: . ::.:: .
GENPEPT_460627 KRRFLAPRHILKD--VVEIANLPDLYKVFERC--
GENPEPT_825572 KRRFLAPRHILKD--VVEIANLPDLYKVFERC--
GENPEPT_7595954 RSHLLPPKHFTEDGNVLQLANLPDLYKVFERC--
GENPEPT_1724118 RSHLLPPKHFTEDGNVLQLANLPDLCKVFERC--
MLH1_HUMAN RSHILPPKHFTEDGNILQLANLPDLYKVFERC--
GENPEPT_3192877 KKYLLPPR---IKDQIYELTNLPTLYKVFERC--
GENPEPT_3880333 KRKFIPPEHFKQAGVIRQLADSHDLYKVFERCGT
: ::.*. : :::: * ******
¡@
3. Perform BOXSHADE program to get a color-coded plot for the
results of question 2.

¡@
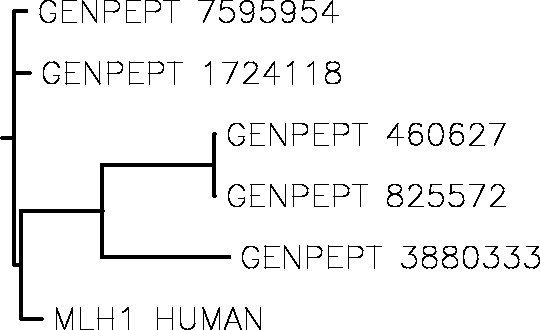
4. Draw rooted phylogenetic tree for these proteins.