1. 1. Search human Mlh1 or its homologous proteins in Protein Data Bank (PDB) database. You just need to search for protein structure, do not find complex structure. Give its (a) ID number, (b) Name of the molecule, (c) experimental method for structure determination and (d) Resolution of the structure.
Get in to the website and go in to the SearchLite in Search the Achieve. Key in the Mlh1 and do the search. There would be shown that 0 structure be found. So we have to key in the MutL. There were two of three that was match:
1B62 Deposited: 11-Jan-1999
Exp. Method: X-ray Diffraction Resolution: 2.10 A
Title Mutl Complexed With ADP
Classification DNA Mismatch Repair
Compound Mol_Id: 1; Molecule: His Tag; Chain: B; Engineered: Yes
Mol_Id: 2; Molecule: Mutl; Chain: A; Fragment: Atpase Fragment; Engineered:
Yes; Biological_Unit: Homodimer
1B63 Deposited: 20-Jan-1999 Exp.
Method: X-ray Diffraction Resolution: 1.90 A
Title Mutl Complexed With Adpnp
Classification DNA Mismatch Repair
Compound Mol_Id: 1; Molecule: Mutl; Chain: A; Fragment: Atpase Fragment; Engineered:
Yes; Biological_Unit: Homodimer
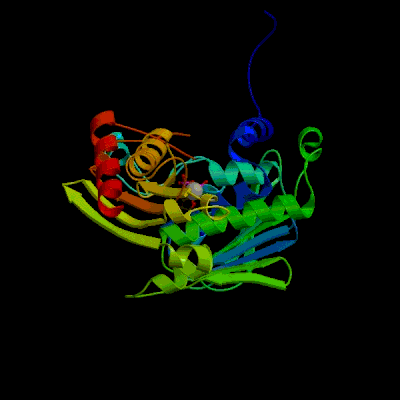
2. Show its structure in ribbons form (400x400).
cilck in the "Explore", and then the View Structure at the left. Key in the size as 400 in the Costom Size Image. Then create image. Here I just took 1B62 Mut Complexed With ADP.

3. Give the structural informations of this PDB file (a) number of protein
chains, (b) number of helix in one protein chain, (c) the first and last residues
in the helix 6 .
In the Sequence Detail, we can find these information:
Overview
Chains Residues Mol. Weight [D] Chain Type
1B62:A 349 38952 Protein
1B62:B 6 822 Protein
Secondary Structure Elements given below are documented in the Help Section
--------------------------------------------------------------------------------
Chain 1B62:A
Compound Mutl
Type Protein
Molecular Weight 38952
Number of Residues 349
Number of Alpha 11 Content of Alpha 30.66
Number of Beta 16 Content of Beta 24.64
Sequence and secondary structure
Showing that the first and last residue in the helix 6
are P-prolin and R-arginine.
--------------------------------------------------------------------------------
Chain 1B62:B
Compound His Tag
Type Protein
Molecular Weight 822
Number of Residues 6
Number of Alpha 0 Content of Alpha 0.00
Number of Beta 0 Content of Beta 0.00
Sequence and secondary structure
1 HHHHHH
4. Give the Class, Fold, Superfamily and Family of this protein in Structural
Classification of Proteins (SCOP).
The SCOP is in the Structural Neighbors. In SCOP, We choose the Protein: DNA mismatch repair protein MutL from Escherichia coli [d.122.1.2]. There would be shown:
Protein: DNA mismatch repair protein MutL from Escherichia
coli
Lineage:
Root: scop
Class: Alpha and beta proteins (a+b)
Mainly antiparallel beta sheets (segregated alpha and beta regions)
Fold: ATPase domain of HSP90 chaperone/DNA topoisomerase
II/histidine kinase
8-stranded mixed beta-sheet; 2 layers: alpha/beta
Superfamily: ATPase domain of HSP90 chaperone/DNA topoisomerase
II/histidine kinase
Family: DNA gyrase/MutL, N-terminal domain
Protein: DNA mismatch repair protein MutL
Species: Escherichia coli
Another one they gave here is:
Protein: DNA mismatch repair protein MutL from Escherichia
coli
Lineage:
Root: scop
Class: Alpha and beta proteins (a+b)
Mainly antiparallel beta sheets (segregated alpha and beta regions)
Fold: Ribosomal protein S5 domain 2-like
core: beta(3)-alpha-beta-alpha; 2 layers: alpha/beta; left-handed crossover
Superfamily: Ribosomal protein S5 domain 2-like
Family: DNA gyrase/MutL, second domain
Protein: DNA mismatch repair protein MutL
Species: Escherichia coli
5. (a) Show Ramachandran plot for this PDB file. (b) How many residues are
in the most favoured regions of Ramachandran plot?
In the Other Sources, there is one PROCHECK under the Analysis kind. Key in the right one 1b62 (in fact, it became failed connection when I try to key in the PROCHECK one) . A new window shows details about Ramachandran Plot statistics and G-factors with a link that make access to the plot. I keep the data here.
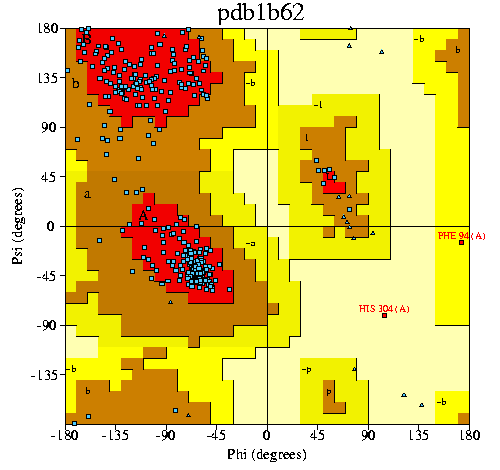
There are 257 residues are in the most favoured regions.