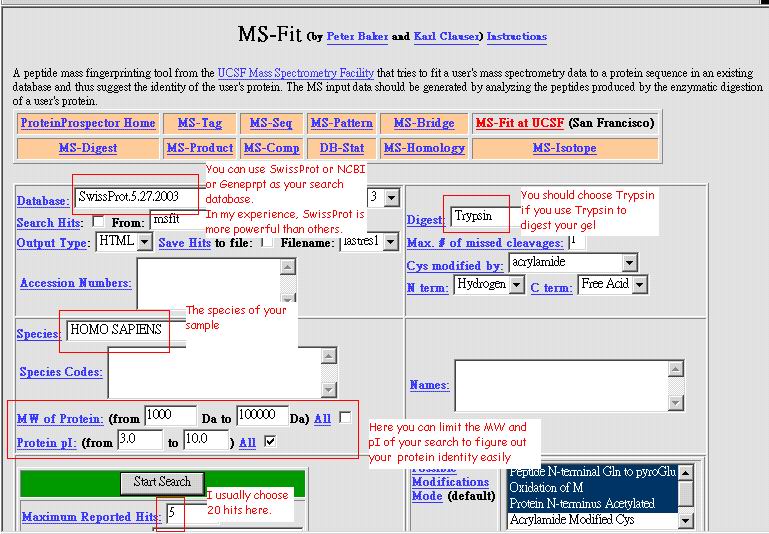
Using
MS-Fit to search possible protein identity in Protein Prospector on UCSF
(http://prospector.ucsf.edu/)
| 1. get your sample
peptide data from IBMS core facility (MALDI-TOF) 2. key in (http://prospector.ucsf.edu/) 3. Choose MS-Fit 4. You can choose your search request as below: |
|
|
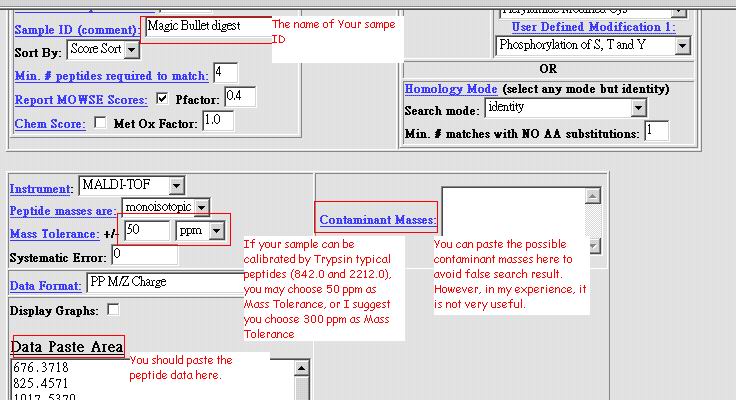 |
| 5. Push the "Start
Search" button 6. Analyze your search result. Here is an example: |
 |
| MOWSE Score:
It is typical to get the score more than 1.0e+004 Masses Matched: The number of alignment peptide/ The percentage of the alignment peptide in total peptides of the protein. Protein MW/ pI : the calculated protein Molecular Weight and pI value. However, the actual value of MW and pI could be different from the calculated values. Proteins could be modified very much. Accession #: The protein identity in Swiss Prot |
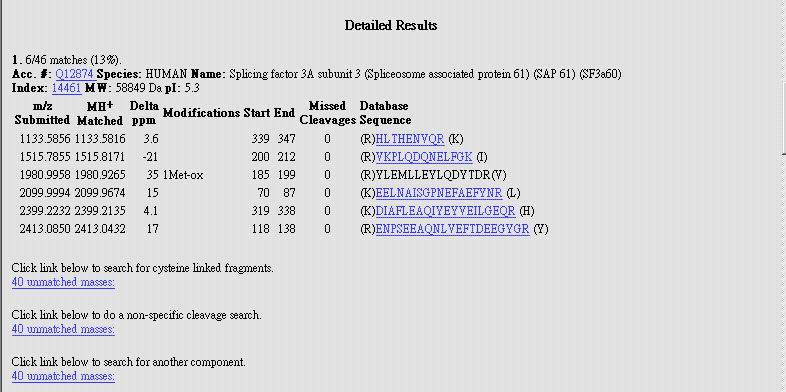 |
|
Detailed Results-> 1. Figure out the difference
between m/z submitted and MH+ Matched to determine the reality of your
search result. |