Homework 8
Sequence analysis / model building
- due on 5/28/98
- David Liu, a student in the department of Lfe Sciences, got a clone from E coli. Here is the DNA sequence:
TGGATGCCATGTTCCGGAGGTAATATGAAGAAATCAATATTATTTATTTTTCTTT
CTGTATTGTCTTTTTCACCTTTCGCTCAGGATGCTAAACCAGTAGAGTCTTCAAA
AGAAAAAATCACACTAGAATCAAAAAAATGTAACATTGCAAAAAAAAGTAATA
AAAGTGGTCCTGAAAGCATGAATAGTAGCAATTACTGCTGTGAATTGTGTTGTA
ATCCTGCTTGTACCGGGTGCTATTAATAATATAAAGGGAACTAAACAGTTCCCT
TTATATTTGTTCTGATTCTGATGATGTCTGTAACGTATGTCCTGTTGCTTTGTTG
AATAAATCGA
Unfortunately, he doesn't know how to use the sequence analysis tools availabled in the internet since he did not take Bioinformatics course before. Could you help him to do the following analysis?
- (1) Find its corresponding polypeptide sequence (DNA -> Protein translation).
- (2) Identify this protein. Is it a new protein?
- (3) Report the total number of negatively charged residues and positively charged residues.
- (4) Color the protein by its charge.
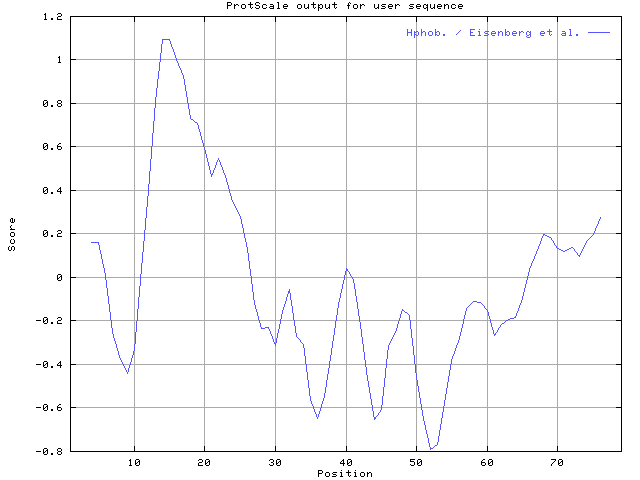
- (5) Draw the hydrophobicity map for this protein using Eisenberg hydrophobicity scale with window size 7. The relative weight of the window edges compared to the window center should set to 40%.
- (6) Please help him to use Prosite scanning tool to find out possible functions or pattern of this protein.
- (7) Calculate its pI and molecular weight.
Answer
1.Using the ExPASy tool menu, we find DNA translate to Protein tool. We type the sequence and find the following message.
W Met P C S G G N Met K K S I L F I F L S V L S F S P F A Q D A K P V E S
S K E K I T L
E S K K C N I A K K S N K S G P E S Met N S S N Y C C E L C C N P A C T G C Y Stop
Stop Y K G N Stop T V P F I F V L I L Met Met S V T Y V L L L C Stop I N R
2.We use BLAST tool of NCBI, and we find the scores of HEAT-STABLE ENTEROTOXIN A4 PRECURSOR are highest. The identities is 100%. Press this to get more information. So, maybe it is not a new protein.
3.Using SAPS tool, we find numbers of negative charge residues (Asp+Glu) are 6 and positive charge residues (Arg+His+Lys) are 12.
4.positive charage(blue):K,R,H
negative charage(red):D.E
polar but unchage(yellow):S,T,C,Y,N,Q
nonpolar(green):G,A,V,L,I,M,F,W,P
MPCSGGNMK KSILFIFLSV LSFSPFAQDA KPVESSKEKI
TLESKKCNIA KKSNKSGPES MNSSNYCCEL
CCNPACTGCY
5. We use ProtScale, and setted the condition of Eisenberg hydrophobicity scale with window size 7and the relative weight of the window edges compared to the window center should set to 40%.

6.We use Prosite scanning tool and find the message.
7.Using computer pI/Mw tool, we find the following message:
pI:8.66
MW:8555.96
¡@