Ubiquitin
Ubiquitin, a small protein ( 8.5kd ) present in all eukaryotic cells . It contains a highly conserved sequence
of 76 amino acids that is identical in a wide variety of sources including humans, fish, and insects. It participates in diverse
cellular functions, such as protein degradation, chromatin structure, and heat shock, by conjugation to other proteins through
the carboxy terminus. This protein is highly conserved in evolution : yeast and human
ubiquitin differ at only 3 of 76 residues. The carboxyl-terminal glycine of ubiquitin becomes covalently
attached to the £`-amino group of lysine residues of proteins destined to be degraded. The energy
for the formation of these isopeptide bonds comes from ATP. Three enzymes- E1 : ubiquitin activating
enzyme°BE2 : ubiquitin-carrier protein°BE3 : ubiquitin-protein ligase- participate in the conjugation of
ubiquitin to protein. First, the terminal carboxylate group of ubiquitin becomes linked to a sulfhydryl
group of E1 by a thioester bond. Activated ubiquitin is then shuttled to a sulfhydryl of E2. Finally, E3
catalyzes the transfer of ubiquitin from E2 to the target protein. A protein tagged for destriction usually
acquires several molecules of ubiquitin. The£`-amino group of a lysine residue of one ubiquitin molecule can become linked to the terminal carboxylate of another. This ubiquitinylated protein is degraded by an ATP-powered 26S protease complex and
ubiquitin is then recycled.

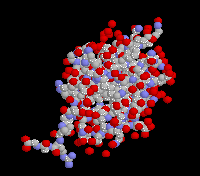
The structure of ubiquitin
The crystal structure of human erythrocytic ubiquitin( right ) has been refined using a restrained least-squares
procedure. A total of 58 water molecules per molecule of ubiquitin are included in the strucyurel. The last four residues in the molecule appear to have partial occupancy or
large thermal motion. The overall structure of ubiquitin is extremely compact and tightly hydrogen-bonded; approximately 87%
of the polypeptide chain is involved in hydrogen-bonded secondary structure. Prominent secondary structural features include
three and one-half turns of alpha-helix, a short piece of 3(10)-helix, a mixed beta-sheet that contains five strands, and seven
reverse turns. There is a marked hydrophobic core formed between the beta-sheet and alpha-helix. The molecule features a
number of unusual secondary structural features, including a parallel G1 beta-bulge, two reverse Asx turns, and a symmetrical
hydrogen-bonding region that involves the two helices and two of the reverse turns. ( From J Mol Biol 194: 531-44 (1987) )
 PDB report
PDB report

 [ Home ]
[ Home ]
 [ Homework]
[ Homework]



 PDB report
PDB report
